Hepatitis B: DNA isolation and analysis method
- Aspirin: Study Finds Greater Benefits for These Colorectal Cancer Patients
- Cancer Can Occur Without Genetic Mutations?
- Statins Lower Blood Lipids: How Long is a Course?
- Warning: Smartwatch Blood Sugar Measurement Deemed Dangerous
- Mifepristone: A Safe and Effective Abortion Option Amidst Controversy
- Asbestos Detected in Buildings Damaged in Ukraine: Analyzed by Japanese Company
Hepatitis B: DNA isolation and analysis method
Hepatitis B: DNA isolation and analysis method. It is suitable for the analysis of a large number of samples and is suitable for medium and high throughput screening.
A DNA preparation method suitable for high-throughput PCR quantification of hepatitis B virus genome. On August 24, 2020, Korean researchers published the paper in the scientific journal Viruses. According to the researchers, in order to establish a simple, economical, and suitable for standard q PCR analysis of HBV DNA preparation method, we designed this column-free DNA preparation method (see figure below: from Viruses).
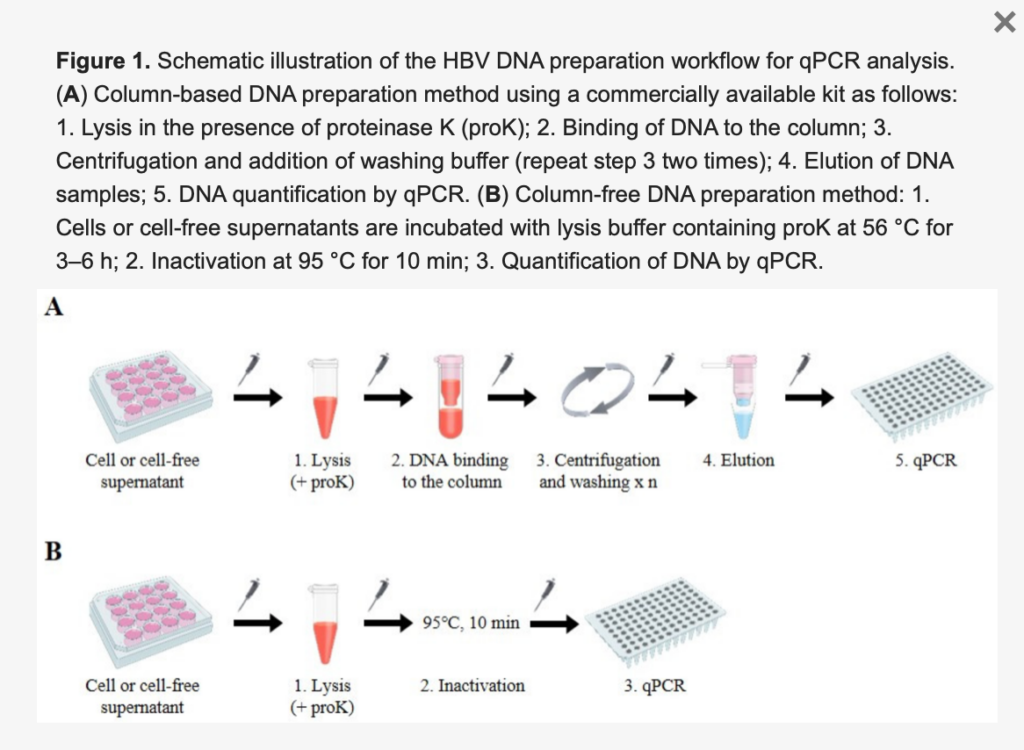
Hepatitis B DNA separation and analysis method, for a large number of sample analysis, and suitable for high-throughput screening
Previously, there have been roughly this new preparation method. According to the researchers, the quantification of the hepatitis B virus genome is usually carried out in the laboratory to evaluate hepatitis B virus replication and antiviral efficacy. Researchers improve time and cost efficiency by eliminating multiple washing and centrifugation steps. Therefore, the material cost and the number of pipetting steps can be reduced, and the throughput of analyzing samples with 96-well or 384-well microtiter plates can be improved. Therefore, it is currently possible to prepare HBV DNA for up to 384 individual HBV cell culture samples.
And accurately quantify the hepatitis B virus genome inside and outside the cell. Another method for researchers to provide comparable sample throughput per unit time requires cost-intensive automated robotic technology, which is unaffordable by most research laboratories, and is usually only used for diagnostics and other highly specialized laboratories. In addition, compared with column-based DNA purification kits, our column-free DNA preparation method requires fewer pipettes, columns, tubes, and buffers for each sample, which significantly improves the environmental footprint.
More importantly, although the column-free method developed by Korean researchers is simple, the quality/purity of DNA prepared using this method is suitable for standard sequence analysis, and its read length is about 500 base pairs, with very low background. At the same time, the column-free method is suitable for preparing hepatitis B virus DNA from the serum samples of chronic hepatitis B virus infection. The data reproducibility of these two DNA preparation methods can show the point. The results show that extracting part of the double-stranded viral DNA genome from hepatitis B virus particles and digesting the capsid with a mixture of detergent and proteinase K can obtain DNA of sufficient purity and use it for accurate qPCR analysis and sequencing.
In this study, the qPCR protocol used can tolerate this purity of DNA and therefore does not require laborious and expensive column-based purification procedures. Even from a single well of a 384-well plate, the PCR template abundance of the recovered HBV genome is low, and the measurement window, detection stability and repeatability are high, as shown by the average %CV and Z’values of 2.2 and 0.63, respectively . These characteristics make this method suitable for high-throughput DNA quantification, such as screening new hepatitis B virus replication inhibitors and DNA secretion containing viral ions.
In this study, Korean researchers demonstrated that it is possible to prepare column-free DNA from a single well of a 384-well plate, and this method is suitable for antiviral drug testing because the LMV EC50 value is comparable regardless of the plate format of. In contrast, column-based DNA purification kits are not suitable for medium and high throughput methods and have the smallest and largest DNA yields. Therefore, samples with too low or too high DNA abundance cannot be recovered completely, and the limitation of sample recovery rate may lead to inaccurate PCR quantitative results.
In layman’s terms, Korean researchers reported the successful development of a new and simple DNA preparation method, which is mainly suitable for parallel preparation of a large number of samples. This method provides high-quality DNA suitable for qPCR and sequence analysis. The new method does not require DNA binding or washing steps, and only requires a short centrifugation step to remove samples after proteinase K treatment. In addition, Korean researchers infer that this column-free DNA preparation method can be applied to other DNA viruses, and may also be applicable to RNA viruses.
Xiaofan Health Conclusion: The above scientific research opinions and conclusions come from the laboratory researchers of the Applied Molecular Virology Laboratory of the Pasteur Institute in South Korea, including Eunji Jo, Jaewon Yang, Alexander Koenig, Seung Kew Yoon, Marc P. Windisch, and will be in 2020 Published in the scientific journal “Viruses” on August 24, 2005. According to the researchers, we have developed a HBVDNA separation and analysis method, which can be fast, simple, low-cost, environmentally friendly, and quantitatively analyze a large number of samples at the same time, and is suitable for medium and high throughput screening.
(source:internet, reference only)
Disclaimer of medicaltrend.org