A new method for colon cancer prognosis based on spatial proteomics
- Statins Lower Blood Lipids: How Long is a Course?
- Warning: Smartwatch Blood Sugar Measurement Deemed Dangerous
- Mifepristone: A Safe and Effective Abortion Option Amidst Controversy
- Asbestos Detected in Buildings Damaged in Ukraine: Analyzed by Japanese Company
- New Ocrevus Subcutaneous Injection Therapy Shows Promising Results in Multiple Sclerosis Treatmen
- Dutch Man Infected with COVID-19 for 613 Days Dies: Accumulating Over 50 Virus Mutations
Cell DIVETM application: Establish a new method for colon cancer prognosis based on spatial proteomics
- Red Yeast Rice Scare Grips Japan: Over 114 Hospitalized and 5 Deaths
- Long COVID Brain Fog: Blood-Brain Barrier Damage and Persistent Inflammation
- FDA has mandated a top-level black box warning for all marketed CAR-T therapies
- Can people with high blood pressure eat peanuts?
- What is the difference between dopamine and dobutamine?
- What is the difference between Atorvastatin and Rosuvastatin?
- How long can the patient live after heart stent surgery?
A new method for colon cancer prognosis based on spatial proteomics.
The birth of new technologies often makes some contributions to the development of the industry.
Today, we interpret the research paper titled “Spatial domain analysis predicts risk of colorectal cancer recurrence and infers associated tumor microenvironment networks” published by users of the University of Pittsburgh Cell DIVETM in the journal Nature Communication[1] to get a feel for the Leica Cell DIVETM Give colon cancer prognosis estimating method innovation “pushing back force”.
Research Background
Colorectal cancer (Colorectal Cancer, CRC) is the third largest type of cancer in the world, and accounts for the second largest number of cancer-related deaths (Figure 1) [2].
Although CRC patients are staged and treated by the mature TNM classification, the heterogeneity of the tumor causes the prognosis of colon cancer patients with the same classification to be quite different.
Statistics show that a considerable proportion of CRC patients have cancer recurrence and metastasis 1-2 years after surgery, and their lives and health are endangered [2].
Active review and consolidation therapy after surgery can avoid the risk of colon cancer recurrence.
With the deepening of CRC research, researchers have found that the heterogeneity of CRC is reflected in the tumor microenvironment and is related to the prognosis of patients [3].
In response to this discovery, people use tumor molecular phenotype, tumor microenvironment composition and tumor T cell infiltration and other indicators to establish colon cancer prognosis prediction and judgment methods [4].
In recent years, some methods for judging the prognosis of colon cancer have been proposed (such as Immunoscore®, etc.).
However, the existing prognostic inference methods are mostly based on traditional pathological techniques and are limited by the number of labeled antibodies in a single test.
They cannot achieve comprehensive detection with multiple indicators and spatial information, which leads to a bottleneck in the development of CRC prognostic inference methods.
The development of multi-label immunohistochemistry technology has helped us break through this limitation.
Today we are following the author of this article is to use Leica Cell DIVETM ultra-multi-standard tissue imaging analysis technology to achieve a new method of CRC prognosis inference-the establishment of computational analysis and systems biology platform (analytics computational and systems biology platform, SpAn platform) .
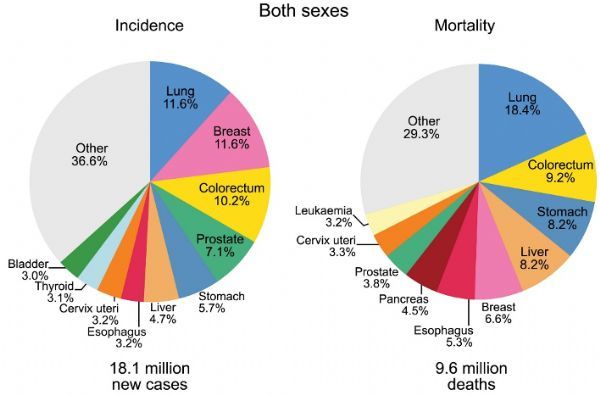 Figure 1 Statistics of the proportion of new cases and deaths of different types of cancer (2018)[2]
Figure 1 Statistics of the proportion of new cases and deaths of different types of cancer (2018)[2]
Research Brief
The author of the article paid attention to the potential of its predecessor technology (MultiOmyx technology) in CRC microenvironment detection during the development of Cell DIVETM, and immediately used it for the development of CRC prognosis inference methods after Cell DIVETM was launched.
With the help of Cell DIVETM, the author easily completed the antibody selection link in the experimental design.
The author selected 55 biomarkers from the library of more than 400 kinds of antibodies certified by Cell DIVETM as detection targets: including common epithelial, immune and stromal cell lineages and classification marker molecules, as well as signaling pathways related to CRC typing Key proteins, molecular markers related to extracellular transport and metabolism, functional proteins related to tumor suppression, biomarkers related to oncogenes, cell adhesion related proteins, molecular markers related to cell and matrix structure, cell types and their status Related characteristic molecules, etc.
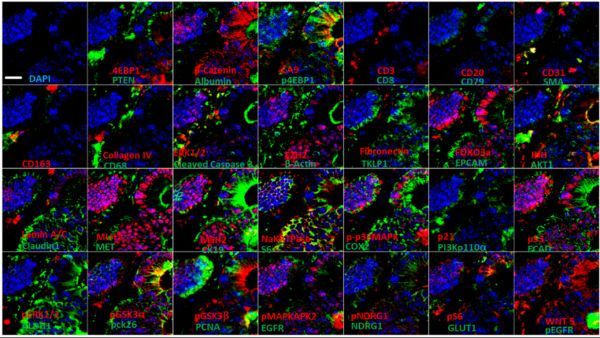
Figure 2 Ultra-multi-label fluorescence histochemical imaging of colon cancer tissue (ruler, 100 μm)
Subsequently, the author used the Cell DIVETM imager to perform ultra-multi-standard fluorescence immunohistochemical imaging of the tissue microarray samples of 432 patients with TNM stage I to III CRC collected by the Clearview Cancer Institute in the United States (Figure 2), and used Cell DIVETM supporting analysis software completes the automatic identification of different tumor regions (Figure 3).
According to tissue structure-specific marker molecules, the tumor microenvironment is divided into epithelial area, mesenchymal area and epithelial-stromal area (the 100 μm interstitial and malignant epithelial cell interaction area at the junction of epithelium and mesenchyme).
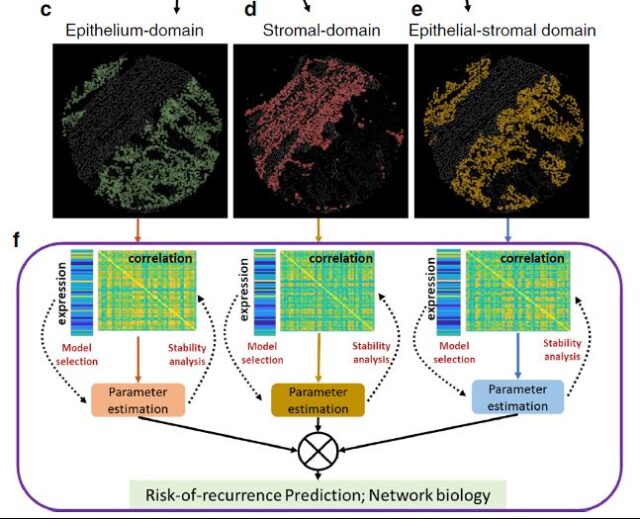
Figure 3 Auto-recognition of colon cancer tissue area and correlation analysis of markers
After the automatic partitioning is completed, the author expands the correlation analysis of the 55 detected markers, and finds a series of prognostic markers from them through recursive operations (Figure 4).
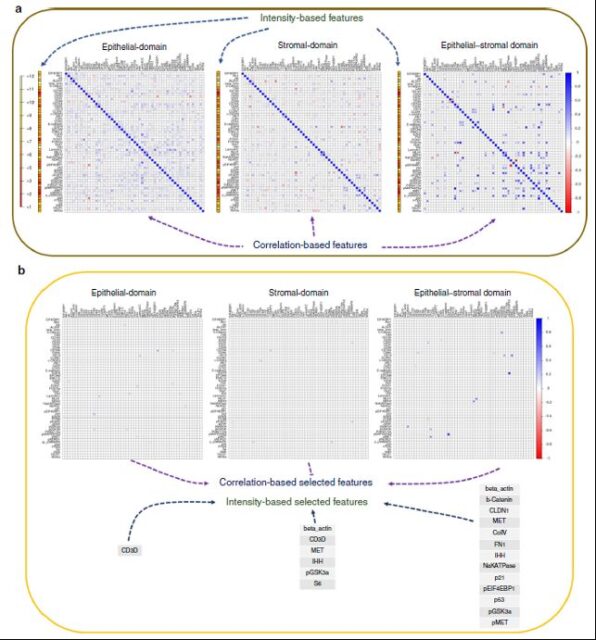 Figure 4 Analysis of regional specific markers of SpAn tumor microenvironment
Figure 4 Analysis of regional specific markers of SpAn tumor microenvironment
Next, the author uses artificial intelligence learning to combine the expression pattern of marker molecules with the spatial information of the tumor microenvironment, and develops the SpAn platform of the CRC prognosis inference calculation model, so as to successfully divide the CRC patients into the postoperative high recurrence risk group and the low recurrence risk group (Figure 5).
Unlike previous methods for CRC prognosis inference, the SpAn platform does not require additional input of patient phenotype data, and prognosis can be inferred based on the spatial proteomics information of the tumor microenvironment.
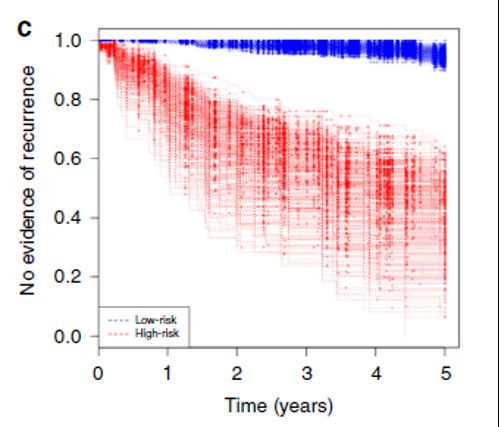
Figure 5 5-year CRC recurrence risk survival curve test on SpAn platform
Finally, the comparison between the SpAn platform and the existing CRC prognostic inference methods shows that the average AUROC of SpAn predicting the risk of recurrence of CRC patients in 5 years is 88.5% (SE=0.1), which is significantly better than the current state-of-the-art methods.
It is believed that in the near future, the SpAn platform can help medical workers accurately formulate postoperative consolidation treatment plans for CRC patients.
The Cell DIVETM ultra-multi-standard tissue imaging and analysis overall solution, from upstream sample preparation, to staining and imaging, to downstream data analysis, can provide a complete set of experimental solutions, breaking through traditional H&E staining, immunohistochemistry and traditional immunofluorescence10 Within the limit of one biomarkers, the qualitative and quantitative analysis of more than 60 biomarkers on a tumor tissue slice can be realized, and the spatial information mining of the tumor microenvironment at the single-cell level can help the research of tumor immunotherapy.
(source:internet, reference only)
Disclaimer of medicaltrend.org
Important Note: The information provided is for informational purposes only and should not be considered as medical advice.