Predicting drug response of EGFR mutation in non-small cell lung cancer
- Did Cloud Seeding Unleash a Deluge in Dubai?
- Scientists Identify Gut Bacteria and Metabolites that Lower Diabetes Risk
- OpenAI’s Model Matches Doctors in Assessing Eye Conditions
- UK: A Smoke-Free Generation by Banning Sales to Those Born After 2009
- Deadly Mutation: A New Monkeypox Variant Emerges in the DRC
- EPA Announces First-Ever Regulation for “Forever Chemicals” in Drinking Water
Predicting drug response of EGFR mutation in non-small cell lung cancer based on machine model
- Red Yeast Rice Scare Grips Japan: Over 114 Hospitalized and 5 Deaths
- Long COVID Brain Fog: Blood-Brain Barrier Damage and Persistent Inflammation
- FDA has mandated a top-level black box warning for all marketed CAR-T therapies
- Can people with high blood pressure eat peanuts?
- What is the difference between dopamine and dobutamine?
- How long can the patient live after heart stent surgery?
Predicting drug response of EGFR mutation in non-small cell lung cancer based on machine model
Around the world, about 1.6 million people die from lung cancer each year, and 85% of lung cancers have a common tissue phenotype, which is called non-small cell lung cancer (NSCLC).
The activating mutation of epidermal growth factor receptor (EGFR) is the most common driving factor of NSCLS, and it often occurs in the kinase domain (exons 18-21) of EGFR.
Although targeted therapies for common EGFR mutations and a small number of other mutations have been approved for marketing, there are still some EGFR mutants without effective treatment options, and the incidence of these mutations and drug sensitivity are still unclear.
The Heymach team at the Anderson Cancer Center of the University of Texas in the United States divided EGFR mutations into four different subgroups based on the structure-function relationship between EGFR mutations and drug sensitivity, which can predict the prognostic effect of patient inhibitor treatment.
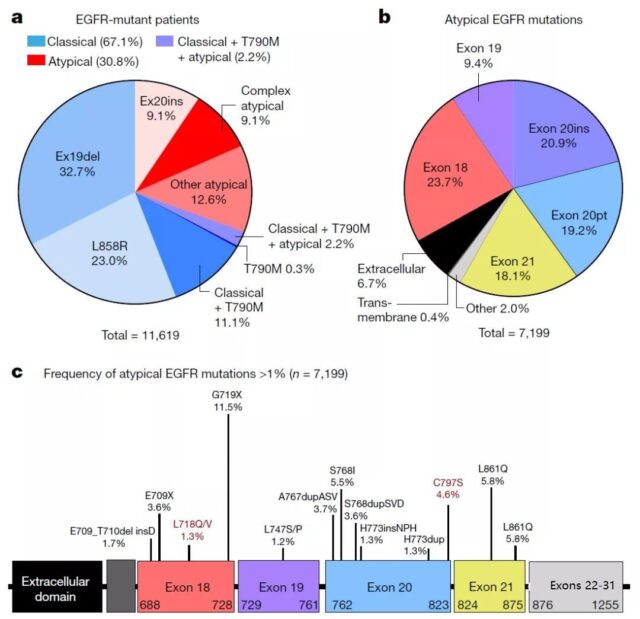
As shown in the figure above, they collected the mutation frequency data of 16,715 NSCLC patients with EGFR mutations in 5 databases, of which 11,619 patients had genotype data. Among these patients, 67.1% carried classic mutations (L858R and or Ex19del with or without T790M), and 30.8% carried atypical mutations.
Among the 7199 atypical mutations with a mutation frequency greater than 1%, the most mutations occurred in exons 20 (40.1%) and 18 (23.7%), mainly in the P-loop region (L718) of the kinase domain. –V726, 13.6%) and the C-terminal of αC-helix (A767–G779, 29.4%).
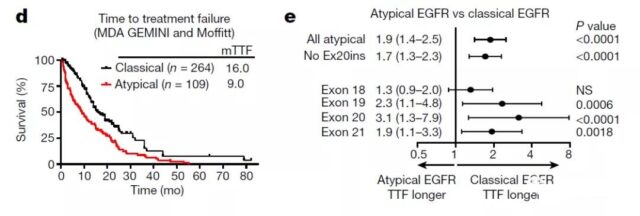
Time to treatment failure (TTF) statistics for patients with different genotypes show that after treatment with different EGFR tyrosine kinase inhibitors (TKIs), the survival time of patients with atypical mutations is significantly shorter than that of classics Patients with mutations.
This shows that clinically, patients with atypical mutations have poor prognosis because there is no effective treatment.
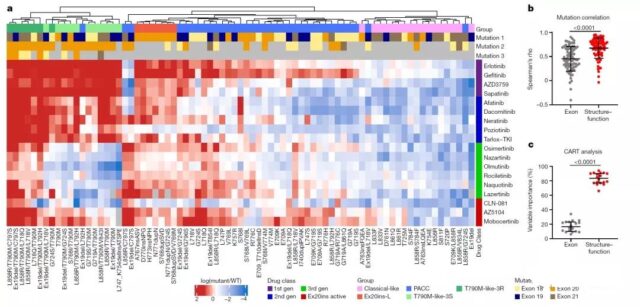
They next analyzed 76 cell lines expressing EGFR mutations in exons 18-21, and screened these cell lines using 18 EGFR inhibitors. After hierarchical clustering, the sensitivity of EGFR mutations to the corresponding inhibitors was obtained as shown above. Sex spectrum.
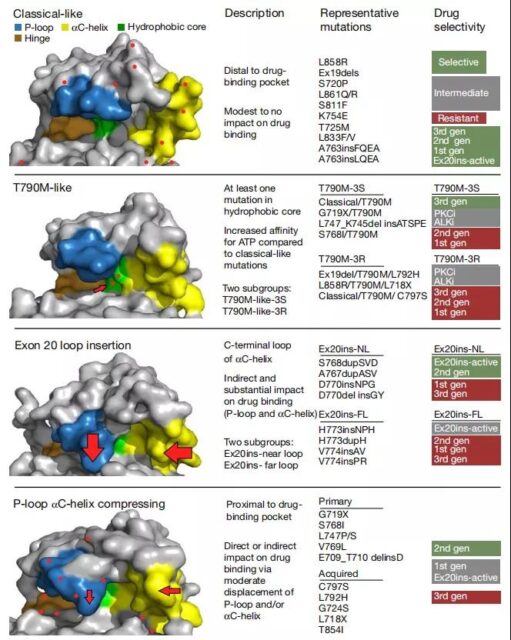
They divided the mutations into four different subgroups based on the mutation’s sensitivity to the drug and the effect of the mutation on the kinase structure. The four different subgroups are as follows:
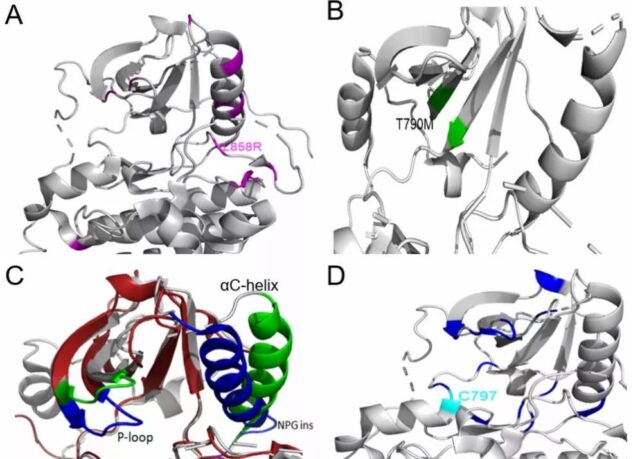
(A) classical-like mutations away from the ATP binding pocket;
(B) t790m-like mutations in the hydrophobic core (T790M-like);
(C) exon 20 Insertion in the loop region of the c-end of the αC-helix (Ex20ins-L);
(D) Mutations in the inner surface of the ATP binding pocket or the c-end of the αC-helix may cause the deformation of the αC-helix or the P-loop region (P-loop and αC -helix compressing, PACC).
Sensitivity spectroscopy shows that the performance of the mutation group based on structural function classification is better than that based on exon classification.
In order to verify this hypothesis, they used Spearman’s method to test the correlation between the two grouping variables and mutation sensitivity, and used the machine learning method of Classification and Regression Trees (CART) to analyze the effects of the two grouping variables on the drug.
Choose importance in decision-making. The analysis results found that classification based on structure and function can predict the sensitivity of mutants to drugs more effectively than based on exons.
Next, the researchers verified the drug sensitivity of the four types of mutants in various aspects.
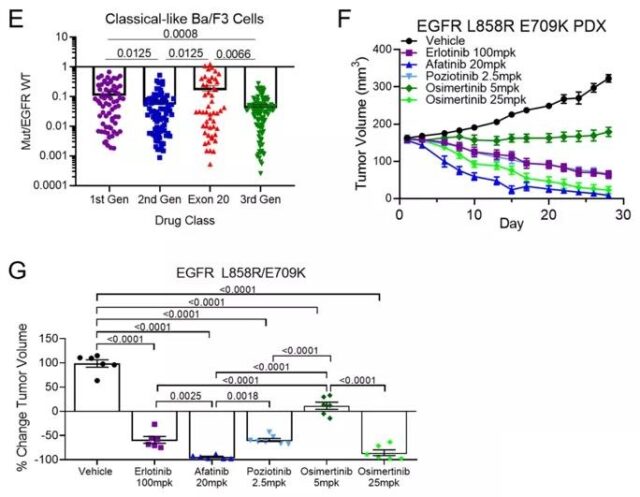
Compared with wild-type EGFR, classic and atypical EGFR mutations have little effect on the overall structure of EGFR. It is sensitive and selective to all kinds of EGFR-TKIs, especially the third-generation TKIs.
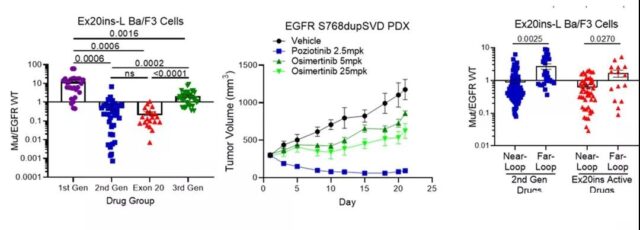
Previous studies have shown that the response of exon 20 mutations to TKIs is heterogeneous. Classification based on structure and function: most of the 20th exon point mutations are PACC mutations; the 20th exon insertion in the αC-helix is a classic-like mutation; the 20th exon insertion in the c-terminal loop of the αC-helix is a unique Subgroup (Ex20ins-L). In vivo, Ex20ins-L is only sensitive to second-generation TKIs and Ex20ins active TKIs.
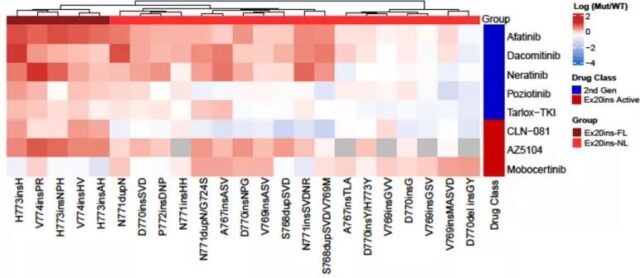
Even in the Ex20ins-L mutation, a certain degree of heterogeneity in drug sensitivity was observed. Compared with the far loop insertion (Ex20ins-Far-Loop), the near loop insertion (Ex20in-Near-Loop) is more sensitive to the second generation and Ex20ins active TKIs.
These data indicate that mutations within a single exon are highly heterogeneous, and exon-based classification is unlikely to provide the best guidance for clinical treatment decisions.
T790M-like mutants carry more than one hydrophobic center mutation, and there are two different subgroups-third-generation TKI-sensitive mutations (T790M-like-3S) and third-generation TKI-resistant mutations (T790M-like-3R) .
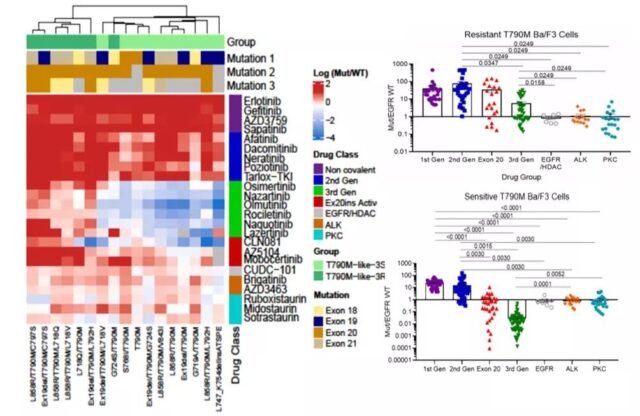
Previous reports have shown that protein kinase C (PKC) and anaplastic lymphoma kinase (ALK) inhibitors have off-target activity against EGFR mutations including T790M. The T790M-like-3S mutant has high selectivity for third-generation TKIs and some Ex20ins activity inhibitors, and moderate selectivity for ALK and PKC inhibitors.
T790M-like-3R mutant contains complex mutations of T790M And a known resistance mutation (ie C797S, L718X or L792H), resistant to EGFR TKIs, but reserved for some ALK and PKC inhibitors such as brigatinib or midostaurin sex.
Through larger-scale screening of PKC and ALK inhibitors, it is possible to develop new covalent inhibitors against T790M-like-3R mutants.
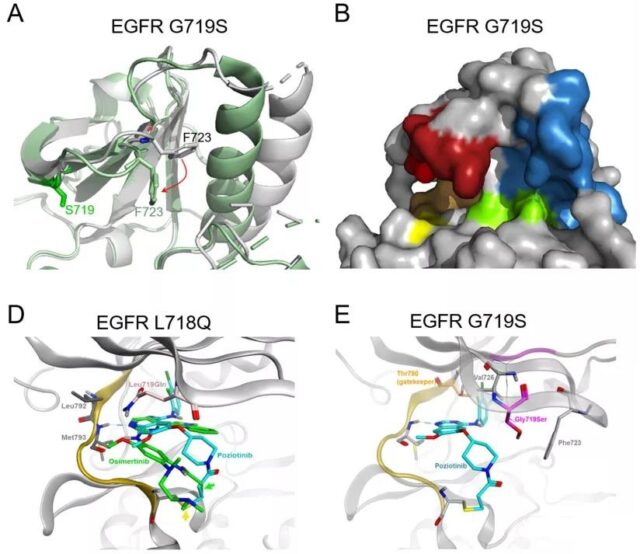
PACC mutations are distributed in each exon 18-21, including G719X, L747X, S768I, L792X and T854I, and are predicted to change the orientation of P-loop or αC-helix.
As shown in the figure above, the simulation of the interaction between osimertinib and PACC mutations G719S and L718Q found that the rotation of the P-loop changes the position of the TKI stable point, making the inert ring of osimertinib away from the P-loop , The combination is not stable.
In contrast, the second-generation TKIs (pozitinib) do not interact with the P-loop of EGFR, but act on the hydrophobic cleft of EGFR.
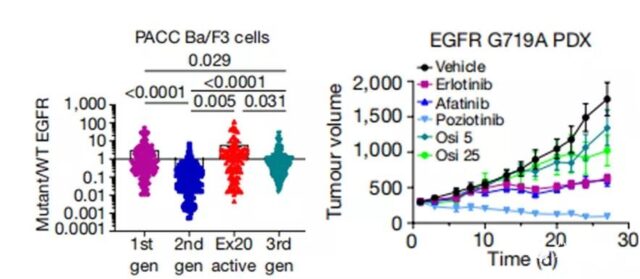
In in vivo experiments, the PDX model derived from patients with G719A mutant NSCLC is resistant to the third-generation TKI osimertinib, but is most sensitive to the second-generation TKI pozitinib.
The above data combined with the PACC mutant sensitivity map shows that the selectivity of the second-generation TKIs to PACC mutations is significantly higher than that of any other TKIs.
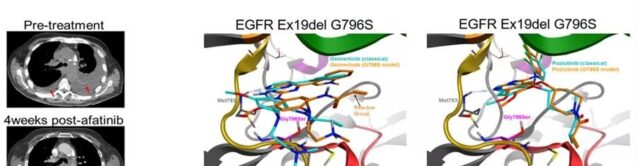
It is worth noting that a patient with a complex PACC mutation (E709K+G719S), the tumor shrank after treatment with the second-generation TKIs afatinib.
The simulation of the acquired PACC mutant showed that the hinge region of EGFR shifted, which made osimertinib bind to M793 unstablely, and the acrylamide group was far away from C797.
The changes in the hinge area did not affect the binding of the second-generation inhibitors.
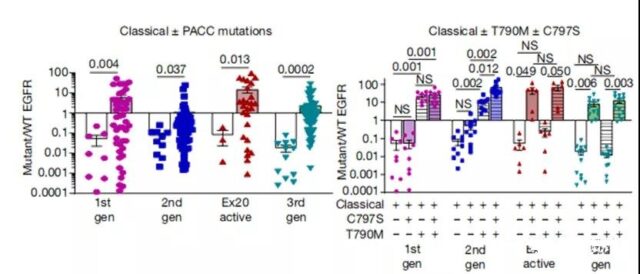
Previous studies have reported that even in the absence of T790M, the C797S mutation will produce resistance to third-generation TKIs. Further in vitro drug screening also showed that the occurrence of PACC mutation has no effect on the resistance of the third-generation inhibitor and the occurrence of T790M mutation has no effect on the resistance of the second-generation inhibitor.
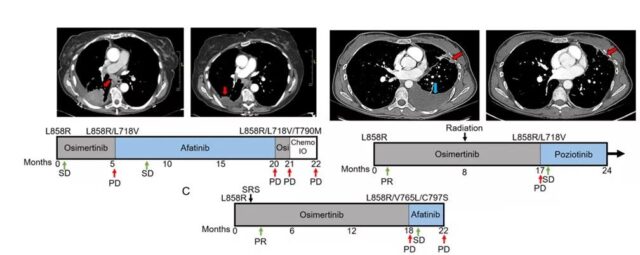
Furthermore, their retrospective study found that 3 NSCLC patients with EGFR L858R mutation developed EGFR-dependent drug resistance after receiving osimertinib for a period of time.
PACC mutations were found in the biopsies at the advanced stage of drug resistance, and then they received second-generation TKI treatment, and the tumor entered the stable stage.
These data indicate that both primary and acquired PACC mutations are sensitive to second-generation TKIs. Grouping based on structure and function can identify a new type of PACC mutations.
The use of second-generation TKIs for this type of mutation is more sensitive than third-generation TKIs. High selectivity and efficacy.
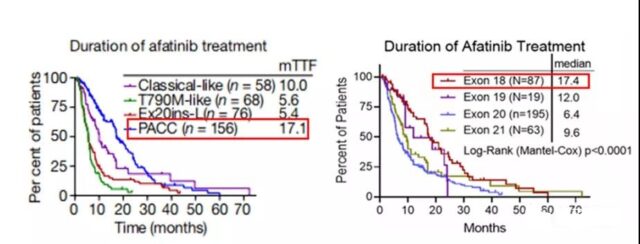
In order to verify whether the classification based on structure and function can better identify the treatment plan with better treatment effect for patients compared with classification based on exons. They used a public database to record the clinical efficacy of patients with atypical EGFR mutations treated with afatinib.
The group based on structure and function found that the duration of treatment (DOT) of PACC mutation patients (n=156) was significantly longer than that of the subgroup (DOT: 17.1 months, P<0.0001).
Based on the exon grouping structure, patients with mutations in exon 18 (n=87) had longer DOT than patients with mutations in exons 19-21 (DOT: 17.4 months, P<0.0001). This group of patients The number is about half of the PACC mutant group.
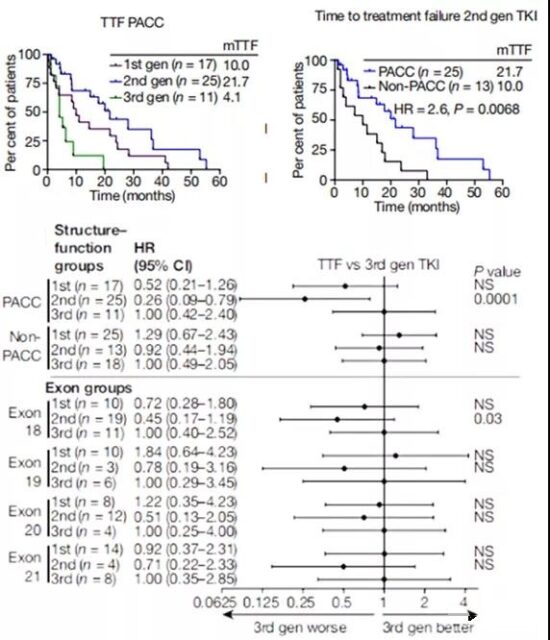
The analysis also found that based on the structure and function grouping, the TTF of PACC mutant patients treated with second-generation TKIs was significantly longer than that of patients treated with third-generation TKIs. In contrast, in patients with non-PACC mutations, there was no significant difference in TTF under different EGFR TKIs treatments.
In addition, when receiving second-generation TKIs, patients with PACC mutations have longer TTFs than patients with non-PACC mutations. When the patients were stratified by exons, the TTF difference between the second-generation and third-generation TKIs was only observed in patients with exon 18 mutations, and the P value was much greater than that of the PACC group.
Therefore, the classification based on structure and function not only identifies a larger patient subgroup, but also this group is treated with second-generation TKIs with better clinical effects.
Summary
this study found that EGFR mutations including atypical mutations can be divided into four different subgroups according to function and structure, and groups based on structure and function and taxa based on exons are useful in predicting drug sensitivity and The phenotype of the disease is better.
This reflects that mutations in different gene regions may have similar effects on protein structure.
Combined with the classification framework provided by this study, comprehensive second-generation sequencing for NSCLC patients can allow clinicians to more efficiently perform personalized EGFR TKIs therapy.
Finally, these findings support the idea that for cancers containing different mutations in oncogenes, the use of a structure-function-based approach may provide meaningful guidance for clinical trial design and drug development.
references
1. Robichaux, Jacqulyne P et al. Structure-based classification predicts drug response in EGFR-mutant NSCLC. Nature, 10.1038/s41586-021-03898-1. 15 Sep. 2021, doi:10.1038/s41586-021-03898-1
2. Drilon, Alexander et al. A Next-Generation TRK Kinase Inhibitor Overcomes Acquired Resistance to Prior TRK Kinase Inhibition in Patients with TRK Fusion-Positive Solid Tumors. Cancer discovery vol. 7,9 (2017): 963-972. doi: 10.1158/2159-8290.CD-17-0507
3. Gonzalvez, Francois et al. Mobocertinib (TAK-788): A Targeted Inhibitor of EGFR Exon 20 Insertion Mutants in Non-Small Cell Lung Cancer. Cancer discovery vol. 11,7 (2021): 1672-1687. doi:10.1158 /2159-8290.CD-20-1683
Predicting drug response of EGFR mutation in non-small cell lung cancer based on machine model
(source:internet, reference only)
Disclaimer of medicaltrend.org
Important Note: The information provided is for informational purposes only and should not be considered as medical advice.