Nano-sequencing “chases” the COVID-19 change
- Normal Liver Cells Found to Promote Cancer Metastasis to the Liver
- Nearly 80% Complete Remission: Breakthrough in ADC Anti-Tumor Treatment
- Vaccination Against Common Diseases May Prevent Dementia!
- New Alzheimer’s Disease (AD) Diagnosis and Staging Criteria
- Breakthrough in Alzheimer’s Disease: New Nasal Spray Halts Cognitive Decline by Targeting Toxic Protein
- Can the Tap Water at the Paris Olympics be Drunk Directly?
Nano-sequencing “chases” the COVID-19 change
Nano-sequencing “chases” the COVID-19 change. Following the confirmation of a mutated new coronavirus in the United Kingdom on December 14, 2020, many countries have recently announced the presence of confirmed cases of the mutated new coronavirus in their respective countries.
This is an unprecedented feat
Laboratories all over the world are studying viral genome sequence data at an unprecedented speed and sharing it with colleagues around the world at the fastest speed. So far, the Global Influenza Data Sharing Initiative (GISAID) website has collected 550,000 new coronavirus genetic data.
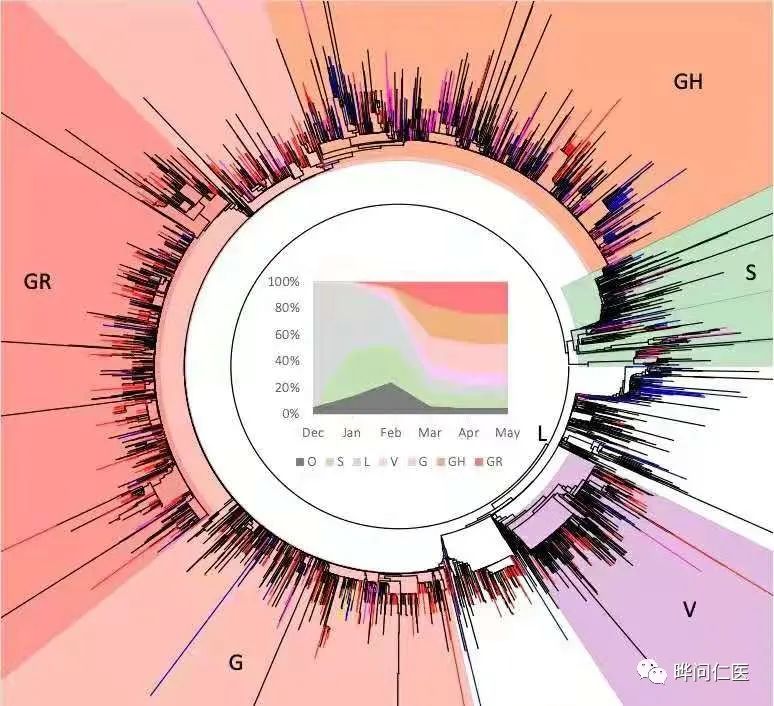
#GISAID is a clade based on the collection of new coronavirus gene data. This clade records mutations in six advanced phylogenetic groups based on the early division of S and L and the further evolution of the virus-L becomes V and G, then G Become GH and GR
Virus evolution is nothing more than constant error replication, which is a random process, a process of constant accumulation. Most errors will not affect the function of the virus. But once a certain necessary mutation appears, the virus can achieve its ultimate goal: invasion, collapse, recombination, shedding, until it invades again, the cycle repeats, this will be an endless host chain.
If the prevention fails, in the end, what we see is that the virus will go out of an overwhelming evolutionary path and find its perfect habitat.
This points to an attractive future: we may be able to read the virus tree faster. So is there a way to quickly track the spread of SARS-CoV-2?
Pioneering rapid genomics method
Thanks to the cutting-edge “nanopore” genome sequencing technology, researchers at the Gavin Institute of Medical Research and the Kirby Institute of the University of New South Wales in Sydney have developed the fastest coronavirus genome sequencing strategy in Australia to date. Technological advances may provide critical and timely clues to the association between SARS-CoV-2 infection cases.
Every time the SARS-CoV-2 virus spreads from person to person, replication errors may occur, which may change several of its 30,000 genetic letters. By identifying this genetic variation, it is possible to identify different cases of the coronavirus and know where it might have spread from.
Genome testing is essential for tracking the spread of the virus, and if you only investigate known epidemiological contacts, you still cannot know its source. By reconstructing the evolutionary history or “family tree” of the virus, it can help to understand the spread of COVID-19 and identify the so-called “super spreaders”. Dr. Ira Deveson, head of the genome technology group at the Garvan Kinghorn Clinical Genomics Center and the corresponding author of the original text, said: “When a new’mysterious’ coronavirus case is discovered, every minute is important. At Garvan, we reuse With the genome sequencing function, the coronavirus genome can be quickly analyzed in just a few hours.”
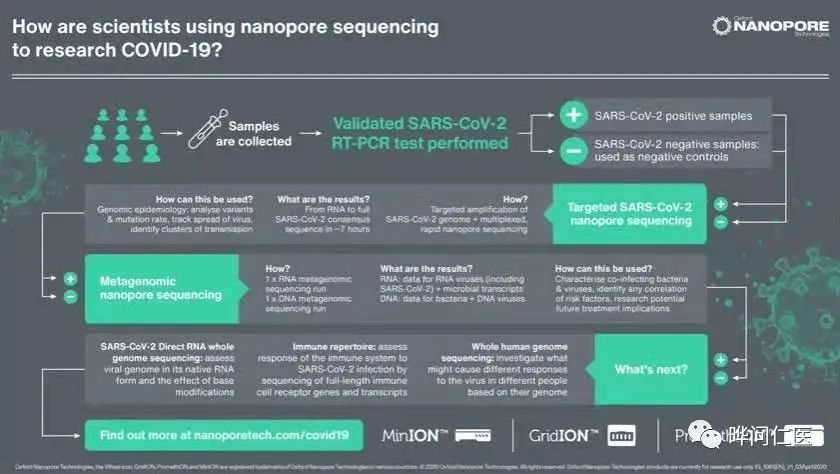
The rise of possible new technologies
High-precision emerging technology. The current gold standard method can only read short gene sequences of 100-150 genetic letters at a time, while nanopore technology has no upper limit on the length of DNA fragments that can be sequenced, and can quickly determine a complete viral genome sequence.
However, like many emerging technologies, people are also concerned about the accuracy of nanopore sequencing. In the paper, the author solved these problems, and reported the results of strict analysis and evaluation of the coronavirus genome sequencing program.
The researcher’s analysis shows that the nanopore sequencing method has high accuracy (sensitivity of variants detected in 157 SARS-CoV-2 positive patient specimens> 99%, accuracy> 99%), and provides the most The researchers hope that these guidelines will promote the adoption of the technology by other teams around the world.
Dr. Deveson said that nanopore sequencing can even enhance SARS-CoV-2 surveillance by enabling point-of-care sequencing and shortening the turnaround time of critical cases. Nanopore devices are cheaper, faster, and more portable, and do not require the laboratory infrastructure required by current standard pathogen genomics tools.
The research published in Nature Communications was supported by the UNSW COVID-19 Rapid Response Research Program, the Medical Research Future Fund (researcher grant APP1173594), the New South Wales Cancer Institute and the Kim Horn Foundation.
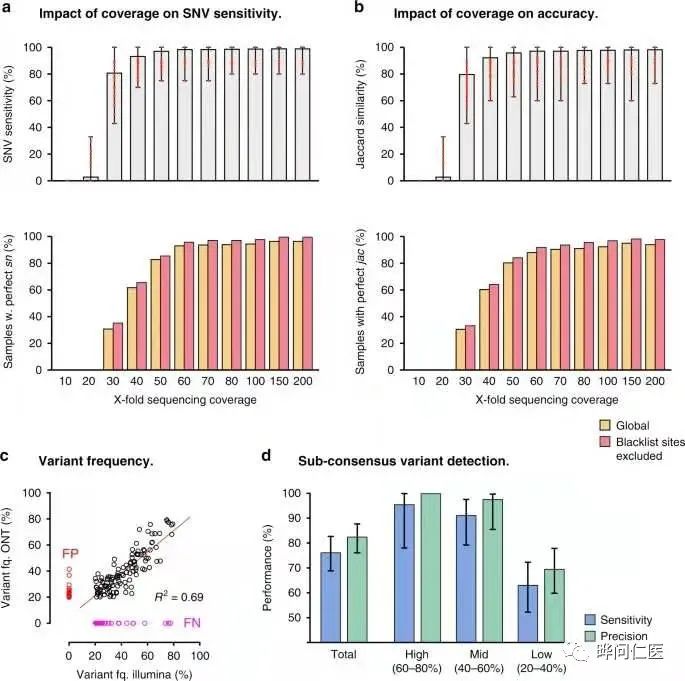
The mutation of the virus is a mechanical adjustment based on the “fault” of the virus itself, and humans should make progress through reflection.
(source:internet, reference only)
Disclaimer of medicaltrend.org



