Improve the performance of foot-and-mouth disease vaccine
- Normal Liver Cells Found to Promote Cancer Metastasis to the Liver
- Nearly 80% Complete Remission: Breakthrough in ADC Anti-Tumor Treatment
- Vaccination Against Common Diseases May Prevent Dementia!
- New Alzheimer’s Disease (AD) Diagnosis and Staging Criteria
- Breakthrough in Alzheimer’s Disease: New Nasal Spray Halts Cognitive Decline by Targeting Toxic Protein
- Can the Tap Water at the Paris Olympics be Drunk Directly?
Improve the performance of foot-and-mouth disease vaccine
Improve the performance of foot-and-mouth disease vaccine. Abstract: Foot-and-mouth disease is still one of the most infectious diseases in the livestock industry, especially from an economic point of view. Seven types of viruses have been identified, of which six different types of foot-and-mouth disease viruses continue to spread throughout the world. As the virus mutates, the virus of each serotype also has diversity.
In addition to matching the serotype of the virus, the vaccine also needs to match the strain of the outbreak virus to ensure its protection. Vaccination has played an important role in helping Europe to eradicate foot-and-mouth disease, but vaccination work will only be initiated in the event of a major outbreak. Therefore, once the virus is introduced accidentally (or deliberately), European animal populations are as susceptible to the virus as North American animals, so the European region will produce nearly 3 billion doses of vaccine each year to control overseas epidemics.
The current vaccine production is made by chemical inactivation after the virus is cultured and amplified in a sterile environment. This type of vaccine can effectively prevent foot-and-mouth disease, but the duration of immunity is quite limited (about 6 months), and vaccination cannot provide sterile immunity or delay the development of disease in susceptible animals. In addition, this type of vaccine is very unstable and usually requires cold chain transportation and storage. In some epidemic areas of the world, it is difficult to ensure preservation of cold chain transportation.
How to develop better vaccines has aroused great interest among researchers, and a variety of methods have been used to make significant progress in vaccine development. However, there is currently no alternative vaccine that can be produced commercially. Globalized disease control is undoubtedly beneficial to all countries, because it reduces the risk of the virus spreading to disease-free areas.
Preface
Foot-and-mouth disease (FMD) is still one of the most worried infectious animal diseases in countries with highly developed animal husbandry industries, as described in the review [1-3]. The disease is caused by infection with foot-and-mouth disease virus (FMDV), a member of the picornavirus family. The virus infects many animal husbandry animals such as cattle, pigs, sheep, goats, water buffalo and more than 70 species of cloven-hoofed animals.
The diagnosis of the disease is mainly based on clinical symptoms, including fever, excessive salivation, blisters around the mouth and inside the mouth, blisters on the coronary artery belt between the fingers and feet, etc. Blisters may also appear on the nipples of female animals. Of course, similar clinical symptoms may also be caused by other viral infections. Therefore, in some countries without FMDV infection, laboratory tests (such as using PT-PCR) must be performed on suspicious cases.
Infected animals lose weight, which is likely to cause secondary infections and the disease may lead to a long-term reduction in the production of agricultural and sideline products, such as milk production. In an infected farm, because the virus is easily transmitted between animals, a large part of the animals are often infected, but the mortality rate is very low. Most of the deaths of young animals are mainly caused by myocarditis. After the acute infection period of cattle, buffalo and sheep (except pigs), a large proportion of animals (for example, about 50% of cattle) will be persistently infected by low-level infectious viruses present in the oropharynx.
These animals are called “carriers” in the literature [4-6]. This virulent state is defined as the virus that will exist in the host for more than 28 days after infection. Some animal populations may carry the virus for several months (sheep) or many years (cattle or dairy cows). The epidemiological significance of the virus-carrying animals is controversial; experiments have proved that the virus cannot be transmitted naturally, directly, and linked from the cattle to the cubs.
However, there are reports of cases of transmission from buffalo to cattle [7]. Recently, it has been reported in the literature that the virus is directly transferred from the oral and nasal fluid of the carrying cattle to the oropharynx of the young to cause infection [8]. Therefore, even if the risk of virus carriers is low, it seems to be one of the ways the virus spreads.
Although foot-and-mouth disease virus does not exist in some parts of the world, such as Europe and North America, it is found in many other countries. Africa and South Asia have become prominent, and the disease situation in South America has improved considerably.
There was an outbreak in Venezuela in 2013 and Colombia in 2017. There is no other clinical report of the disease except for this disease [9], but countries in South America are still undergoing large-scale vaccination, which may make the actual situation of virus transmission fail to highlight. It is estimated that the annual direct or indirect economic loss caused by foot-and-mouth disease in disease-endemic countries is approximately US$822 billion [10]. The spread of FMDV to countries that are normally free of the virus may also have huge economic consequences.
It is worth mentioning that the foot-and-mouth disease outbreak in the United Kingdom in 2001 caused a loss of approximately US$10 billion in the country[11]. This epidemic that lasted for more than 8 months affected approximately 2,000 farms and resulted in more than 6 million animals. The death of animals. The disease also spread from the UK to Ireland, France and the Netherlands.
From the 1950s to the 1960s, there was an outbreak of foot-and-mouth disease every year in Europe[12]. Fortunately, the veterinary service was organized in an orderly manner, coupled with widespread vaccination in some countries, and the epidemic was effectively controlled. By the 1970s, the disease was in Europe. The outbreak rate has dropped to a very low level.
Therefore, the European Union has stopped the foot-and-mouth disease vaccine in the 1990s, unless there is an emergency [13]. One consequence of this is that the European animal populations are now susceptible to the disease, but this measure has also greatly promoted the trade of European livestock and their meat products.
The United Kingdom has never used a vaccine before, and even the foot-and-mouth disease outbreak broke out in 2001; in the Netherlands, vaccination is used as a control measure to control the spread of the disease, but all vaccinated animals have to be destroyed subsequently [14].
There are currently 7 different serotypes of foot-and-mouth disease virus known, namely O, A, C, South Africa (SAT) 1,2,3 and Asia-1. Since 2004, there has been no relevant information anywhere. Disease report of type C FMDV.
Therefore, the wild type of this serotype may now be extinct [15]. There is almost no cross protection between different serotypes. Therefore, animals that have been infected or vaccinated with one serotype are still highly susceptible to other serotypes. In fact, because the virus has specificity even in a single serotype, animals vaccinated with a specific virus strain may not be immune to another virus of the same serotype.
Therefore, if vaccination is to be carried out, it is necessary to match the vaccine against the local virus strain that causes the disease, not just at the serotype level. Type O is the most frequently reported virus serotype. According to reports, it was reported that about 70% of global outbreaks were caused by it about 20 years ago [16] and this situation has remained largely unchanged since then [15].
In summary, there are great differences between various serotypes, especially the differences between various viruses from different sources in the world. In addition to the Asia-1 serotype, each virus serotype has been classified into different subtypes according to nucleotide sequence analysis and different geographical distributions [16,17].
The existence of SAT and Asia-1 viruses (as the name suggests) is strictly limited in terms of geographic location, but occasionally they appear beyond their usual areas. In fact, Asia-1 FMDV has appeared in Greece in recent years [18], In Egypt, SAT 2 FMDV also appeared [15]. In contrast, types O and A (previously called serotype C) have a wide geographic distribution.
Document Retrieval
This review is mainly based on the public literature provided in PubMed (http://www.ncbi.nlm.nih.gov/pubmed) and the author’s rich experience on the subject.
Current status of foot-and-mouth disease vaccine
At present, about 2.5 billion doses of foot-and-mouth disease vaccine are used every year in the world, mainly in China and South America[10]. At present, foot-and-mouth disease vaccines are usually infected by virus-infected BHK cells and then cultured in suspension under aseptic conditions to proliferate and use binary ethylene. Imine (BEI) is made by chemically inactivating virus particles. BEI can modify viral RNA and then purify non-structural viral proteins. Before administration, the vaccine is mixed with an adjuvant (to form an oil or water dosage form containing aluminum hydroxide and saponin).
FMDV particles are approximately spherical (about 25-30nm in diameter) and have a protein shell. The capsid is composed of 60 VP1, VP2, VP3, and VP4 molecules, which enclose a single positive-stranded RNA molecule (Figure 1). The RNA genome of FMDV includes a large open reading frame that can act as a messenger RNA to encode a single polyprotein (Figure 1).
But in fact, the complete polyprotein has never been detected, because during and after translation, the polypeptide chain is mainly cleaved by virus-encoded proteases (L and 3C) to generate a variety of primary precursors (P1-2A, P2 and P3). ), which is further processed to collect 15 mature viral proteins (see below). The four structural proteins (VP1-VP4) are produced by P1-2A precursors, while the non-structural proteins (NSP), which are mainly involved in protein processing, RNA replication, and anti-host defense mechanisms, are derived from P2 and P3 precursors.
Virus particle assembly
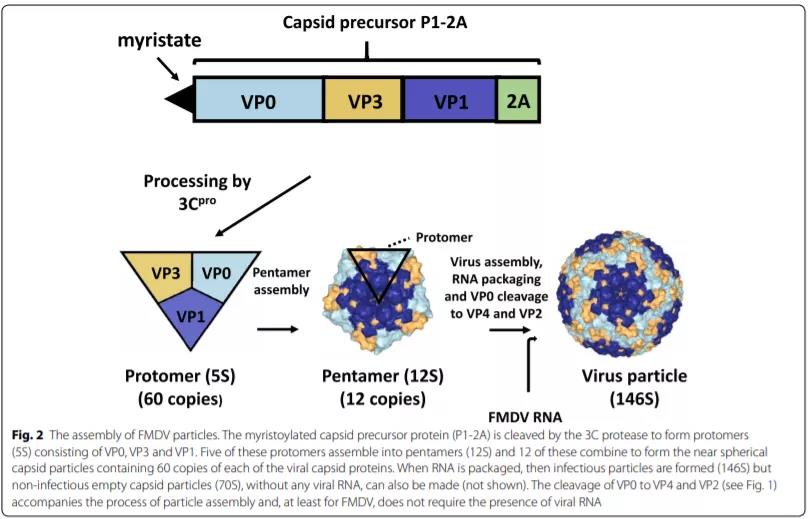
FMDV particles need to be assembled through a series of steps. Various intermediates were identified from the sedimentation characteristics (S value) during sucrose gradient centrifugation. However, because the intermediate must remain stable during the analysis, this analytical method may limit the detectability. The capsid precursor P1-2A (Figure 2) is modified by adding a myristate (C14) group at the N-terminus through the action of the cellular myristoylation system [19, 20], and can also be modified by 3C protease (3Cpro) Cleavage into VP0, VP3, VP1 and 2A peptides, the latter is usually not integrated into the virus, but if the cleavage linking group is modified, it can also be integrated like VP1-2A [21].
Every 5 5S protoplasms are then assembled into 12S pentamers, and twelve pentamers are assembled with the viral genome to form a complete virus particle (146S). In the final assembly step, VP0 is cleaved into VP4 and VP2 by an unknown mechanism. The polymer is assembled into non-infectious empty capsid particles (70S). This process may also occur in the absence of RNA genome. At least for FMDV, the cleavage of VPO seems to depend on the assembly process of the particles, rather than the presence of viral RNA [22, 23], so the assembled empty capsid particles have been found to contain VP2.
The production efficiency of empty capsid particles seems to vary between virus strains. Serotype A empty capsid particles are easily formed in cells infected with FMDV [24], but for most serotype O viruses, empty capsid particles are not sufficient. This may affect the efficiency of using recombinant protein expression systems to generate empty capsid particles of different strains/serotypes [25].
Intact virus particles (or empty capsid particles) are more immunogenic than individual virus components or even partially assembled virus particles (such as pentamers) or their breakdown products [26]. Therefore, it is important to ensure that vaccines contain high levels of intact particles when they are produced and are stored under conditions that can maintain this state (including the use of cold chains in vaccination campaigns). Systems for the detection and quantification of intact particles (certain serotypes) have been developed [27, 28].
Limitations of existing hand, foot and mouth disease vaccines
In summary, the facts prove that vaccines, together with other control measures, including animal activity control, are very effective for disease control in Europe. But the current foot-and-mouth disease vaccine still has shortcomings. Vaccination can prevent the occurrence of diseases, but limited virus replication still occurs in these animals, especially in the oropharynx, which is enough to make those vaccinated animals that are indeed infected become carriers and carry infectious for a long time. virus.
If the virus particles are purified by non-structural proteins (NSPs) during the production process, then serology can be used to distinguish whether an infected animal is vaccinated or infected with a natural virus (DIVA concept). Both viral infection and inactivated vaccines can induce the production of antibodies against the capsid protein. However, anti-NSPs antibodies are only produced by infection (although multiple inoculations with purified vaccines may also cause anti-NSPs responses).
There are currently a variety of detection methods used to detect anti-NSPs antibodies [29]. However, if the vaccinated animals are infected (although there is no disease), even if they become carriers of infectious viruses, the level of virus replication is not sufficient to stimulate the body to produce an immune response against NSPs [29-30].
Some limitations of the currently used foot-and-mouth disease vaccines are discussed further below. Some of the defects are due to the nature of the vaccine itself, and some are due to the biological characteristics of the virus, especially the rapid evolution of the virus, including its gradual genetic drift and more thorough recombination events.
Biological characteristics of FMDV
When FMDV genomic RNA is positive and introduced into the cytoplasm, infection may occur [31-32]. The RNA sequence contains a single, large open reading frame (about 7000 nucleotides (nt)), encoding a long (about 1300 nt) 5’untranslated region (UTR) and a much shorter 3’fan-shaped Polyprotein-UTR (approximately 90nt), followed by poly(A) tail (Figure 1).
Initially, genomic RNA must play the same role as mRNA. It encodes a polyprotein of about 2300 amino acids. However, it does not directly form a complete polyprotein, but is cut into a variety of precursors during and after synthesis, and the precursors are further processed into 15 different mature proteins (Lab, Lb, VP4, VP2, VP3, VP1, 2A, 2B, 2C, 3A, 3B1, 3B2, 3B3, 3C and 3D) (see Figure 1). The above mature protein contains 4 different structural proteins (VP1, VP2, VP3 and VP4) that form the viral capsid (Figure 2).
When the virus is outside the cell, this protein shell (each particle has 60 copies of each capsid protein) can protect the viral RNA and help deliver the viral RNA to the cytoplasm of the new cell, thus starting a new round Infection. The capsid binds to the integrin receptor on the cell surface [33]. Accompanying the invasion of the virus, mild acidification occurs in the cell [34] to destroy the acid-labile viral capsid and release viral RNA, and then begin translation.
Eleven non-structural proteins include two different forms of lead proteases (Lab and Lb), which initiate translational synthesis by two different AUG codons [35] and three different forms of 3B (3B1, 3B2, and 3B3). These three short peptides are also called VPg (linked to the viral protein genome) because they are covalently linked to all recent 5′-ends of synthetic viral RNA (Figure 1). Each VPg can be used as a uracilated substrate to form VPgpUpU[36], and then used as a primer for RNA synthesis.
Among FMDV polyproteins, 3C protease (3Cpro) is responsible for most of the protein hydrolysis process, and 3D protein is RNA-dependent RNA polymerase (3Dpol). The characteristics and functions of different virus-encoded proteins have been reviewed in detail [37, 38], so I will not repeat them here.
Virus RNA replication is error-prone
In addition to being translated as mRNA, the FMDV genome can be used as a template for replication. Therefore, there must be at least one molecule in each infected cell, which cannot replicate to produce accessory strand RNA due to translation activities.
It should be noted that in the process of protein synthesis, ribosomes move along the RNA from the 5’end to the 3’end, while during RNA replication, RNA polymerase synthesizes antisense RNA from the 3’end of the RNA. ; The process of translation and replication cannot occur simultaneously on the same molecule. Then, the newly synthesized negative strand can be used as a template to replicate positive-strand RNA, which can be used for translation (synthesis of viral proteins), or used as a template to replicate more secondary-strand RNA, or can be packaged into new viral particles ( figure 2).
The replication of viral RNA occurs in the structure of the cell’s inner membrane [39], called the replication complex or replication organelle, which contains a variety of host proteins and viral proteins (including 2C and 3Dpol) required for this process [40]. The wrong sequence will be inserted into the RNA replication and the replication of viral RNA is extremely prone to errors. The evaluation of the error rate of RNA polymerase found that, on average, for every 10,000 nt synthesized, approximately one error occurs [41]. Because the synthesis of a new genomic RNA molecule (after copying both the positive and negative strands) must replicate approximately 17,000 nt, this means that almost every FMDV genome will have at least one error.
Picornaviruses have no known proofreading mechanism. Therefore, the RNA sequence of all viruses is equivalent to a closely related sequence library, which is also called the virus quasispecies library [42]. The modification of the 3Dpol fidelity (increase or decrease the error rate) by the picornavirus reduces the “adaptability” of the virus [43]. Therefore, for these viruses, there needs to be a balance between the need to maintain a fully functional RNA sequence and the need to quickly adapt to the new environment.
This continuous replication error has caused the virus population to continue to evolve. However, it should be noted that when some error-fixing errors appear (that is, they become major errors), the “common” sequence of the virus population (that is, corresponding to the major bases at each position in the genome) will only change relatively slowly (and the error Rate comparison), which is probably because they are given certain selection advantages.
The consensus sequence of the FMDV population changes by 0.5-1.0% of the genome every year [44], which means that there will be about 40-80 nt every year or about 1-2 nt every week. Differences in these sequences may change the biological characteristics of the virus (for example, its antigenicity or replication speed), but it can also be used to track the spread of the virus during outbreaks [44-45].
It is foreseeable that based on the analysis of different virus isolates that broke out in the UK in 2001, it can be found that the RNA polymerase error will be in the entire virus genome. The mutation starts from a single source and spreads among unvaccinated animals [44]. The identification of nearly 200 different nt substitution sets in 23 different full-length viral sequences found that about 28 changes were in the non-coding regions (about 14%, close to the total number of these non-coding regions in the genome). proportion).
In the entire coding region of the polyprotein, most of the changes observed are synonymous, and only 40 (20%) of the changes modify the coding protein sequence. Obviously, the amino acid sequence retention is highly selective. The amino acid changes that alter the function of the protein will generally be harmful to the virus itself and therefore will not be maintained. However, some regions of the genome are more tolerant of changes than others. What is certain is that among the immunized population, there will be a selective pressure for different virus strains, and this selective pressure will cause the virus strain to change its antigenicity.
FMDV sequence diversity
Carrillo et al. compared seven serotype representatives of more than 100 FMDV strains [46]. It was found that within the 5′-UTR, the average base identity between all serotypes exceeded 80%, and in the entire polyprotein coding region (about 7000 nt), the sequence identity level between any two virus isolates was at least Is 73%. However, the variability of the VP1 coding region (approximately 639 nt) between strains is greater than most polyprotein coding sequences, and only about 50–70% nt identity between all serotypes [47].
Among the different products derived from polyproteins, VP1 itself has the lowest proportion of invariant amino acids (24%) [46]. It is speculated that this affects the ability of certain residues to accept changes (for example, in the loop region that connects the structural elements within the capsid proteins VP1, VP2, and VP3 that are exposed on the surface) and the immune response to these exposed features of the virus particle. Competitive pressure. The variability of the capsid protein VP4 (located completely inside the intact granule) is much smaller (81% unchanged residues) [46].
Certain surface exposed parts of the capsid protein (including antigenic sites, see below) can vary widely. However, even within the VP1 capsid protein, there are highly conserved gene sequences. For example, RGD (Arg-Gly-Asp) motif is essential for the interaction with cell integrin receptors and for virus attachment and cell entry Is required [33]. Similarly, it is obvious from the comparison conducted by Carrillo et al. that the YCPRP sequence near the C-terminus of FMDV VP1 (Tyr-Cys-Pro-Arg-Pro) is also highly conserved [46], but its importance until recently Just be recognized [48,49].
This gene sequence is necessary for 3Cpro to process the capsid precursor P1-2A, and it is also highly conserved among different picornavirus genera (such as: WCPRP in enterovirus and FCPRP in cardiovirus. Note that W( Trp) and F (Phe) are aromatic amino acids like Y (Tyr)). There is no doubt that the base sequence encoding these conserved amino acids does make mistakes, but it is probably that the resulting virus (if fully feasible) cannot effectively multiply, so these variants will not be retained in the virus population.
RNA recombination
In addition to the gradual accumulation of the aforementioned base sequence changes, more significant genomic evolution involving RNA recombination may also occur in the picornavirus genome. In the process of picornavirus replication, RNA polymerase (3Dpol) may replicate from one positive-strand template to another through a so-called “replication selection” [50] mechanism. This process can lead to the formation of “chimeric” genomes, for example, the capsid coding sequence is derived from a parental virus, while the rest of the genome is derived from different strains of the virus [51, 52].
Therefore, recombination can change the serotype of the virus. Template switching during RNA replication may occur very frequently during RNA replication, but if all genomes in a single cell are closely related, it will not have a significant impact on the results of the RNA replication process and will be difficult to detect . However, if the cell is co-infected with two genetically distinguishable genomes, the recombination may or may not produce a new chimeric genome. If it is produced, it can have different characteristics from each parental virus strain and can replicate preferentially under appropriate conditions.
Obviously, the production and testing of new recombinant viruses requires that animals be infected with distinguishable strains of the virus. In some parts of the world, multiple serotypes of the virus are often co-transmitted, and there are reports of recombination of FMDV serotypes in this area [51, 53, 54] and recombination between different lineages of the same serotype [55]. If a (near) full-length genome sequence is produced, it will facilitate the identification of recombination, and thus the genetic relationship between different parts of the genome and other viruses can be established [54].
How reorganization happens
In summary, the detection of genetic recombination requires animals to be infected with two different FMDV strains at the same time. That is, each virus needs to infect the same cells in the host at the same time so that the viral RNA polymerase can switch between two different FMDV RNA templates. When animals are infected with FMDV, there is usually a relatively short acute infection stage [1, 56]. High levels of viremia appeared within a few days, and blisters 0 lesions containing high levels of virus were observed. As the protective immune response develops and the blisters heal, the infection gradually subsides. However, as mentioned above, many infected animals (about 50% of cattle) cannot completely clear the infection and maintain a low level of poisoning in the oropharynx for months or even years. It should be noted that pigs are not carriers. It is uncertain whether recombination occurs between FMDV when the parental virus causes an acute infection (simultaneous infection in animals or if “carrier” animals with different viruses can co-infect cells with two different virus strains). The extremely high levels of virus present in acutely infected animals may make individual cells in the host more likely to be co-infected, but, obviously, this provides a fairly narrow time frame for cell co-infection. However, reinfection of “carrier” animals with different virus strains may occur within a few weeks, months or even years after the original infection. If the status of the carrier is the key to recombination, then this should not happen to pigs under these conditions. At present, it is not clear in which species the virus recombined, but a recent study proved that recombination occurred between different variants of the experimentally infected African buffalo (Syncerus cafer) FMDV SAT 1 [58]. Therefore, it should be possible to design experiments to determine whether recombination between foot-and-mouth disease viruses will occur between pigs.
Antigen diversity of FMDV and vaccine selection
The existence of 7 serotypes of FMDV clearly shows that the genetic diversity of the virus also leads to the diversity of antigens. The antigenic properties of the virus depend on the surface exposed residues of VP1, VP2 and VP3 (VP4 is completely inside the virus particle [59]). Antigen matching between viruses is important for selecting the most suitable FMDV strain to make a vaccine to prevent disease. In principle, this can be done by conducting a vaccination test: inoculate a potential vaccine in a natural host animal and then challenge it with the current outbreak strain. But this will be very time-consuming and expensive. In actual research, an in vitro neutralization test is usually used to determine whether the antiserum produced by a specific vaccine can effectively neutralize the outbreak strain. But the disadvantage is that the reproducibility of this type of measurement is poor [60].
Vaccine efficacy test
The efficacy test of the FMDV vaccine involves vaccinating a certain number of animals with different doses of the vaccine, and then attacking with an appropriate virus strain (usually homologous) 21 or 28 days after vaccination [61]. Although this method has some advantages, it also has certain limitations in standardization. For example, this method cannot provide information about the vaccine’s ability to prevent non-homologous virus strains or other forms of virus attack, that is, only through the exposure of infected animals, and only involves a single point in time after vaccination, and due to the relatively large number of tested animals Its accuracy is limited.
Paton et al. have published a detailed review of issues related to the evaluation of the protective effects of specific FMD vaccines in the laboratory [60]. There have been some reports that test the effectiveness of FMD vaccine against specific viruses [62]. In addition, a recent review on considering the design and evaluation of vaccine efficacy in this field has been published [63].
FMDV monoclonal antibodies (Mabs) can be used as standards to assess the antigenicity of different viruses. A series of virus-neutralizing monoclonal antibodies of different serotypes were used to select FMDV anti-neutralizing mutants. By sequencing selected neutralization-resistant viruses, the key surface exposed groups of FMDV can be identified, and these residues are critical to the antigenicity of the virus.
Using this method, multiple independent antigenic sites have been identified. Usually, these antigenic sites are located on the surface exposed loops, and each surface exposed capsid protein has a certain effect on antigenicity [64-71]. Therefore, the relationship between the antigenic site defined by the selection of monoclonal antibodies and the epitope recognized by the serum of the natural host animal is not simple. It should be noted that the antigenic sites identified by monoclonal antibodies are indeed consistent with the high sequence variability regions of the strain, indicating that these regions have changed due to competitive pressure.
However, these studies may not find all residues bound by monoclonal antibodies. For example, if the monoclonal antibody binds to a region of the virus that must maintain the viability of the virus (for example, binds to an integrin receptor), then the selected monoclonal antibody resistance mutant will not be modified in that region. In fact, studies have used cryo-electron microscopy to analyze the interaction between a specific monoclonal antibody (D9) and the intact serotype O virus, revealing that the monoclonal antibody and VP1 residue D147 (the RGDL group required for integrin binding) Part of the sequence), which has not been found in previous sequencing of neutralizing resistant mutants [73].
It is shown that the substitution of the adjacent residue L148 in VP1 can lead to the stronger anti-neutralization ability of monoclonal antibody D9 [64,66], so it is not surprising that residue D147 also interacts with the antibody.
In principle, the correspondence between the FMDV outbreak strain and the key antigenic sites of the viral capsid and the known vaccine strain can be evaluated by measuring the reactivity with the corresponding monoclonal antibody or by sequence determination [74-76]. However, the ability of vaccines to protect against viral attacks undoubtedly depends on many factors. There have been analysis reports on the ability of a series of different O-type vaccine strains to resist multiple O-type viruses spreading in southern Asia [77].
Obviously, the overall ability of the vaccine to induce neutralizing antibodies in the host (measured by the in vitro virus neutralization test) is very important, and non-neutralizing antibodies may also reduce circulating virus levels. The strength of the immune response to the vaccine will also depend on the number of antigens in the vaccine and the integrity of its particles. High-potency vaccines can produce protective immunity within a few days [78], and the immune efficacy can last at least 6 months from a single dose [79].
However, the duration of immunity produced by current inactivated vaccines (using standard doses) is usually limited. In endemic areas, it is often necessary to re-vaccinate animals at least twice a year [80] to maintain vaccine protection. Therefore, the time difference between vaccination and exposure to virus attack and the actual level of virus that may be encountered in nature may be important.
It is worth mentioning that some foot-and-mouth disease vaccines, such as the widely used O1 Manisa vaccine, are based on virus strains that have spread widely long ago (O1 Manisa was isolated in 1969). However, despite the genetic diversity of this virus, a highly effective vaccine based on this strain can still provide protection against heterologous serotype O strains [81, 82]. Similarly, despite the low level of antigen “matching”, highly effective serotype A vaccines have demonstrated protection against a variety of serotype A strains [83].
Development prospects of foot-and-mouth disease vaccine
Various current reviews describe the need for improved FMD vaccines [80, 84, 85] and the development status of improved FMD vaccines [38, 86]. Readers can refer to these papers to learn about the main strategies and details of the current development of new FMDV vaccines. In short, only a few major methods have received continued attention. See Figure 3 for details.
Essentially, each method aims to produce virus-like particles that have antigenic sites themselves. Studies have proved that the use of a single capsid protein or synthetic peptide as a new vaccine is unsuccessful. The current focus is mainly on the following three types of vaccines:
(A) The production of non-infectious empty capsid particles by co-expression of myristoylated capsid precursor P1-2A and 3Cpro (Figure 3a) [25, 87, 88].
Viral vector vaccines used to express empty capsid particles in cell culture (such as vaccinia virus or baculovirus). This type of vaccine does not depend on the ability of FMDV particles to cause infection. Therefore, the capsid protein can be modified to enhance assembly. The stability of the particles [88,89]. Such vaccines also provide the potential to produce FMDV antigens without the need for a sterile environment. However, this type of vaccine also has some problems similar to inactivated vaccines, such as short duration of immunity and lack of sterility protection in the carrier animal.
(B) Expression of FMDV empty capsids with replication-deficient viral vectors (Figure 3b) [90,91].
This method is different from the aforementioned non-infective system in that the defective viral vector can infect cells in the host (but will not spread), so FMDV products are produced in the cells of the animal. Compared with the immune response produced by extracellular protein antigens, this type of vaccine can make the host produce a wider range of immune responses.
For example, the human adenovirus vector (Ad5) vaccine has obtained a “conditional license” in the United States. This is the first time that the United States has allowed domestic production of FMDV vaccine. Currently, high-dose vaccines are needed to obtain immune protection, but after about 6 months, the duration of protective immunity against foot-and-mouth disease has decreased [92], while immunity against adenovirus persists. This also restricts the use of this type of vaccine only as an emergency, to fight the daily outbreaks in countries without foot-and-mouth disease.
The alpha virus vector vaccine used by Gull-berg et al. [91] has a very complicated production process, but the advantage is that the alpha virus vector vaccine based on the RNA genome [93] replicates only in the cytoplasm of the cell, similar to the FMDV replication process. Problems related to RNA modification in the nucleus (such as splicing with Ad5 vectors) are not related.
(C) Development of improved FMDV strains:
The strain is attenuated, and the attenuated strain can grow efficiently in cell culture (for example, based on a virus form lacking the Lb coding region (Figure 3c) [94]).
The Lb coding region can be accurately deleted from the FMDV virus, but it does not affect its ability to replicate in BHK cells (eg [94, 95]). These mutant viruses are highly attenuated and cannot easily revert to virulence. Therefore, compared with current vaccine strains, these attenuated FMDV virus antigen sources are safer. A virus leak occurred in time, and the virus was unable to infect animals, causing an epidemic. The production environment requirements for such products are lower than those of conventional FMDV vaccines, but they also have similar characteristics and limitations to conventional vaccines. Like conventional FMD vaccine viruses, the attenuated particles must be chemically treated before use. Inactivate and purify.
Sum up
As people’s knowledge of the biology of FMDV increases, a variety of methods will be developed to design new vaccines. Various vaccines are still being actively developed, but currently only the conventional inactivated vaccines developed in the 1960s can be widely used to combat foot-and-mouth disease. The apparent disappearance of type C foot-and-mouth disease virus worldwide, the significant improvement in the number of mouth-and-mouth disease cases in Europe and the foot-and-mouth disease situation in South America indicate that the current disease control measures are appropriate, including vaccination.
In epidemic areas, it is obviously important to trust existing vaccines. The poor effect of the vaccine may not only be caused by the poor match between the vaccine strain and the epidemic strain, so the quality of the vaccine should be tested during the production process and after storage/transportation.
Many developers are still very interested in developing better foot-and-mouth disease vaccines, because the disease is still a major problem in many countries and may greatly affect the trade of animals and their products in these countries. Global epidemic control is obviously beneficial to all countries, because it reduces the risk of the virus spreading to disease-free areas and causing huge economic losses.
Improve the performance of foot-and-mouth disease vaccine.
Improve the performance of foot-and-mouth disease vaccine.
(sourceinternet, reference only)
Disclaimer of medicaltrend.org



