New Genome Assembly Algorithm – Improving the ability to assemble complete human genomes!
- Aspirin: Study Finds Greater Benefits for These Colorectal Cancer Patients
- Cancer Can Occur Without Genetic Mutations?
- Statins Lower Blood Lipids: How Long is a Course?
- Warning: Smartwatch Blood Sugar Measurement Deemed Dangerous
- Mifepristone: A Safe and Effective Abortion Option Amidst Controversy
- Asbestos Detected in Buildings Damaged in Ukraine: Analyzed by Japanese Company
New Genome Assembly Algorithm – Improving the ability to assemble complete human genomes!
- Red Yeast Rice Scare Grips Japan: Over 114 Hospitalized and 5 Deaths
- Long COVID Brain Fog: Blood-Brain Barrier Damage and Persistent Inflammation
- FDA has mandated a top-level black box warning for all marketed CAR-T therapies
- Can people with high blood pressure eat peanuts?
- What is the difference between dopamine and dobutamine?
- How long can the patient live after heart stent surgery?
New Genome Assembly Algorithm – Improving the ability to assemble complete human genomes!
A new genome assembly algorithm, La Jolla Assembler (LJA), completely reconstructs almost half of the chromosomes in the human genome in a fully automated manner, improving the ability to assemble the complete human genome and reducing error rates.
Compared to other assembly algorithms using HiFi sequencing technology, the error is reduced by a factor of 5. The accuracy of this approach will facilitate large population studies of complex and understudied regions of the human genome.
An international team led by researchers in the UC San Diego Department of Computer Science and Engineering has shown that a new genome assembly algorithm, called La Jolla Assembler (LJA) , greatly improves large-scale genome reconstructions, in which DNA fragments are arranged The process of forming a complete genome is an essential aspect of genome sequencing.
Furthermore, LJA significantly reduced error rates and improved the ability to assemble the complete human genome.
This will make it easier to conduct large-scale population studies, in which thousands or millions of people are sequenced and their genomes compared to better understand the genetic factors that lead to disease.
The study was recently published in the journal Nature Biotechnology under the title ” Multiplex de Bruijn graphs enable genome assembly from long, high-fidelity reads “.
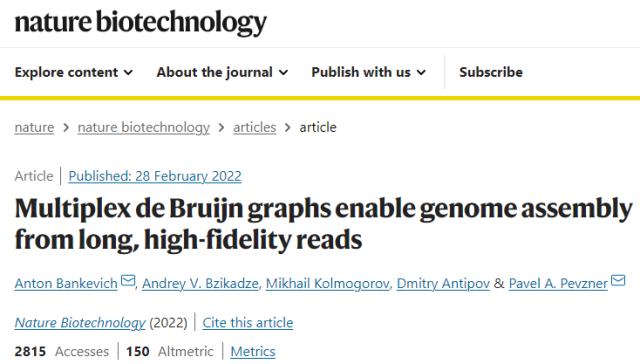
” We used LJA to completely reconstruct almost half of the chromosomes in the human genome in a fully automated fashion ,” said Pavel Pevzner , the paper’s senior author and the Ronald R. Taylor Distinguished Professor of Computer Science . , which reduces assembly errors by a factor of 5. The accuracy of this approach will bring important benefits, especially for large population studies of complex and understudied regions of the human genome, such as centromeres or antibody production sites. “
Genome assembly algorithms are computational tools for reconstructing genomes based on collections of shorter sequences.
For years, researchers have relied almost exclusively on short-read technology, which produces reads of up to 300 nucleotides.
These provide vital genomic information but leave gaps in the genome sequence—many in biomedically important regions. As a result, the Human Genome Project, completed two decades ago, has left thousands of unassembled regions — unexplored DNA that may have clinical and scientific significance.
“This incomplete assembly of the human genome revolutionized biology and medicine 20 years ago,” said lead author Anton Bankevich , a postdoctoral researcher in the Department of Computer Science and Engineering . “However, the missing genome segment may harbor more secret.”
More recently, scientists have begun employing HiFi sequencing technology (over 10,000 nucleotides), which has helped them sequence complete human and other genomes.
The completion of the first complete human genome last year by the Telomere-to-teltelomere (T2T) consortium was an important milestone.
However, this feat requires a lot of work and is nearly impossible to scale to hundreds, let alone millions, of genomes.
To automate the process and improve speed and accuracy, Pevzner’s team employed a computational method called a de Bruijn graph that helps them assemble millions of sequences into complete genomes.
This was originally an obscure mathematical method invented by the Dutch mathematician Nicolaas de Bruijn, which has become a workhorse for sequencing, modeling a genome as a complex network of roads connecting cities (short segments of the genome), finding A method of traversing the network while using each road.
In a sense, this is history repeating itself. More than 20 years ago, Pevzner and colleagues used the de Bruijn graph to understand short readings.
“While it appears that simply applying this 20-year-old technology to HiFi sequencing technology will lead to superb human genome assembly,” said co-author Andrey Bzikadze , a graduate student in the UC San Diego Bioinformatics and Systems Biology Program . But when faced with the extremely complex de Bruijn graph that builds the human genome, all previously developed algorithmic ideas fall apart. Reusing old methods requires a lot of computer memory, making them impossible.”
LJA solves this problem, reducing data footprint and assembly errors.
It lays the foundation for increased speed and accuracy in large-scale population studies, where scientists will need to assemble millions of genomes to identify healthy or disease-causing gene sequences.
“Assembling individual genomes is not enough to drive biological discoveries,” Pevzner said. ” It is by comparing different genomes that scientists can understand their function and associations with disease. That’s why we need to scale up genome assembly efforts and create a The T2T Human Genome produces the same quality genome assembly but can be automated by an algorithm. ”
Reference:
https://phys.org/news/2022-03-highly-accurate-algorithm-scales-ability.html
New Genome Assembly Algorithm – Improving the ability to assemble complete human genomes!
(source:internet, reference only)
Disclaimer of medicaltrend.org
Important Note: The information provided is for informational purposes only and should not be considered as medical advice.