Researchers combine different human antibodies to target SARS-CoV-2
- Why Botulinum Toxin Reigns as One of the Deadliest Poisons?
- FDA Approves Pfizer’s One-Time Gene Therapy for Hemophilia B: $3.5 Million per Dose
- Aspirin: Study Finds Greater Benefits for These Colorectal Cancer Patients
- Cancer Can Occur Without Genetic Mutations?
- Statins Lower Blood Lipids: How Long is a Course?
- Warning: Smartwatch Blood Sugar Measurement Deemed Dangerous
Researchers combine different human antibodies to target SARS-CoV-2
- Red Yeast Rice Scare Grips Japan: Over 114 Hospitalized and 5 Deaths
- Long COVID Brain Fog: Blood-Brain Barrier Damage and Persistent Inflammation
- FDA has mandated a top-level black box warning for all marketed CAR-T therapies
- Can people with high blood pressure eat peanuts?
- What is the difference between dopamine and dobutamine?
- How long can the patient live after heart stent surgery?
Researchers combine different human antibodies to target SARS-CoV-2.
Science: Large-scale research has constructed hundreds of human antibodies binding SARS-CoV-2 profiles, which is expected to develop more effective COVID-19 antibody therapies
In a new study, the researchers released a detailed map of the binding of human antibodies to SARS-CoV-2, which was produced through global collaboration and comparison of almost all leading clinical candidate antibody drugs.
This new research will guide the development of more effective COVID-19 antibody therapies and help scientists develop effective vaccines to deal with emerging virus variants.
Researchers from the La Jolla Institute of Immunology in the United States released a detailed map of the binding of human antibodies to SARS-CoV-2. This map is generated through global collaboration and comparison of almost all leading clinical candidate antibody drugs.
This new research will guide the development of more effective COVID-19 antibody therapies and help scientists develop effective vaccines to deal with emerging virus variants.
The relevant research results were published online in the Science Journal on September 23, 2021. The title of the paper is ” Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: A global consortium study .”

These findings have promoted COVID-19 research in three key aspects:
(1) Classification and binding mapping of hundreds of antibodies provided by more than 50 different organizations around the world; -The exact location of the binding on the spike protein (S protein) of CoV-2;
(2) These researchers describe the neutralization strength, or potency, of each antibody, and the possibility of each antibody to provide protection against virus variants
(3) Antibodies with similar footprints on the S protein are classified as the same “communities”; these researchers have shown how antibodies from different groups can be combined into a powerful antibody “cocktail” to target SARS -CoV-2.
Correspondence paper, La Jolla immunology professor at the Institute, Dr. Erica Ollmann Saphire said, “We were able to draw geographical maps of the S protein, and see which antibodies bind to which of its footprint.
This atlas provides a reference to help predict what Antibodies are still effective against SARS-CoV-2 variants, such as the Delta variant, which has seen a surge in cases.” Saphire led the global effort behind this research called the Coronavirus Immunotherapeutic Consortium (CoVIC).
In fact, these authors discovered that three different sets of antibodies can protect against mutations in the SARS-CoV-2 S protein. These antibodies can target vulnerable parts of the S protein, even if it is mutated. Saphire said, “We now have a framework for choosing a durable antibody cocktail for COVID-19 treatment.”
Collect powerful antibodies
CoVIC includes approximately 370 antibodies from 59 different discovery projects, covering a wide range: from academic laboratories and small biotech companies to large international pharmaceutical companies. These antibody therapies are being evaluated side by side in a standardized laboratory by seven different partner laboratories.
Saphire and his team are determining the high-resolution structures of these antibodies, and are also rapidly generating tools to study the effect of S protein mutations on antibody efficacy.
Dr. Kathryn Hastie, co-first author of the paper and lecturer at the La Jolla Institute of Immunology , said, “CoVIC was established to analyze a large set of monoclonal antibodies with the same footprint.
The original goal was to study the original SARS-CoV-2 It was quickly discovered that the S protein of this virus is constantly evolving. When you are talking about treating someone with a monoclonal antibody, the ability of the S protein to change has a profound effect.”
Dr. Haoyang Li, co-first author of the paper and a postdoctoral researcher at the La Jolla Institute of Immunology, said, “CoVIC contributors used different strategies to find these antibodies.
The breadth of this antibody makes our research more comprehensive than previous studies. , Because previous studies may have only studied the antibodies of a small number of survivors.”
This systematic study of this huge antibody library is a huge project. This study provides a framework for understanding which antibodies are effective (or ineffective) against which SARS-CoV-2 variants on a global scale.
This information will be the key to shrinking this antibody library and to advance further research from it. Saphire said, “Every member of our 25-person laboratory is involved.”
This global effort, coordinated by project manager Dr. Sharon Schendel, includes blindness to a range of clinical drug candidates so that all therapies can be evaluated on a level playing field.
This global work was coordinated by the project manager, Dr. Sharon Schendel, and involved blindly looking at a series of clinical antibody drug candidates so that all drug candidates were evaluated in a level playing field.
Similarly, Professor Bjoern Peters of the La Jolla Institute of Immunology is leading a team to establish the CoVIC database as a public repository for all data collected by CoVIC partner laboratories.
These data are freely accessible to other researchers to compare and contrast antibodies against SARS-CoV-2 S protein. This comprehensive information will help determine which antibodies will become candidates for clinical use.
Looking for an antibody “all star”
A key region on the S protein is called the receptor binding domain (RBD), which is like a protruding rock. Scientists in the Saphire laboratory use mountaineering terms to describe its structure. The interior and exterior of the RBD are connected by a central “valley”. There is a “mountain peak” and a “Pingdingshan” tower next to it. Cross Pingdingshan and look down at the “cliff” below.
By observing where the antibody binds to the RBD, these authors divided promising antibodies into different “populations.” Schendel said, “A population is a group of antibodies that have the same behavior, meaning that they can or cannot bind to the S protein at the same time as other antibodies.”
Antibodies know how to recognize specific virus structures. They saw their target and grabbed it to prevent the virus from infecting cells. But what if their target changes and the area recognized by the antibody changes?
Saphire said, “A SARS-CoV-2 variant may not be discovered until a few weeks after its appearance — this is an endless catch-up game. We need to know which ones in our antibody library will be in the future. It will last for months and years.”
These authors need to find antibodies that target “conserved” sites on RBD. These sites are very important to the life cycle of this virus, and if they mutate, the virus is likely to fail to function. The most effective antibodies are those that target these conserved sites.
In order to find durable antibodies, Hastie constructed a series of S proteins with different point mutations. Their structure reflects those seen in worrying SARS-CoV-2 variants such as Alpha, Gamma, and Delta.
For some antibodies, one or two mutations in the S protein are enough to make them lose their binding target. Hastie said, “We can see which types of antibodies are evaded by which mutations. It’s really useful to know this because we can see these mutations in worrying variants in the real world.”
For example, scientists know. The Beta variant has a mutation called K417N at one site. Hastie said, “We can now identify antibodies that are affected by mutations at this site, but we can also identify antibodies that are still effective against other sites on the S protein of the Beta variant.”
Schendel and Hastie worked closely with Dr. Daniel Bedinger of Carterra, a biotechnology company based in Salt Lake City.
The company developed the LSA instrument and epitope binning analysis software , which is critical to this analysis. Bedinger drew a network diagram that allowed them to compare how more than 250 antibodies bind to SARS-CoV-2.
These network diagrams show which antibodies will bind to the S protein in the presence of each other, in other words, which antibodies will “get along” with other antibodies. Hastie said, “Carterra voluntarily does this work on a free basis. I want to give Daniel and Carterra a thumbs up.”
The goal of these authors is to find the best antibodies from different groups and combine them into an all-star team to conquer this virus.
At the same time, Li led the structural research. They used the cryo-electron microscopy facility of the La Jolla Institute of Immunology to image the structure of the antibody bound to the RBD.
Through Li’s work, he obtained a “hit map” showing where the antibody targets the virus. The hit map shows that there are seven antibody communities that target variable or conserved regions on the S protein.
In addition, compared to traditional studies using Fab versions of antibodies, this new study uses intact IgG to mimic and reveal how these antibodies protect cells from infection. Some IgGs target the SARS-CoV-2 S protein in a bivalent manner, which increases the affinity and potency of these antibodies, while other IgGs tend to inactivate the virus by cross-linking the S proteins together. Li said, “This is new information for the scientific community, and it is a good example of why we need this more detailed antibody analysis.”
Not only did Li lead the structural biology research work, he also ensured the accessibility of these structures—not only in this paper, but also by storing structural information in a public database so that other scientists can access the data. Schendel said, “Li is the driving force behind obtaining these structures. His efforts are true heroes.”
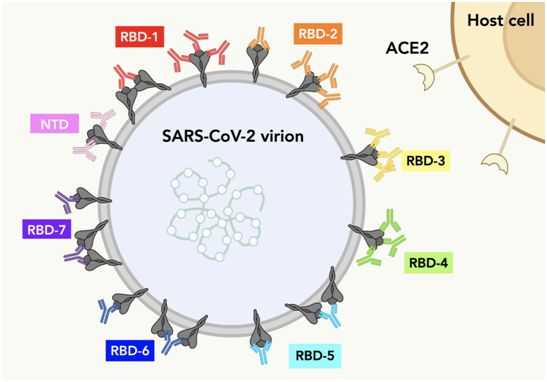
This picture shows a series of spike proteins on the surface of SARS-CoV-2. Each color of antibody represents the possible IgG-S protein binding mode of each RBD population. Picture from Science, 2021, doi:10.1126/science.abh2315.
Finally, these authors assembled a fascinating color-coded antibody library among different antibody populations. This study shows which antibodies can be combined in monoclonal antibody therapy. The study also shows which antibodies need to be elicited from future vaccines to maximize protection from SARS-CoV-2 variants.
Science with global impacts
Three of the antibody populations proved to be particularly durable against RBD mutations. Hastie said, “These antibodies are indeed good candidates for monoclonal antibody therapies. If you are building an antibody cocktail, you will want at least one of these antibodies, because they may remain resistant to most SARS- The efficacy of CoV-2 variants.”
Some COVID-19 patients can already get a cocktail of monoclonal antibodies. As Schendel explained, the current antibody cocktail has limitations. They are best if administered before severe symptoms appear, and they must be administered intravenously in a clinical setting.
Through this new research, CoVIC is closer to developing a more powerful antibody therapy so that it can fight against SARS-CoV-2 variants. Schendel said more powerful antibody therapies may also be effective at lower doses, making them a practical option for distribution in countries with fewer medical services. She hopes that one day, monoclonal antibody therapy can be used as a simple injection.
The CoVIC team is now working with partners on animal protection research. Other CoVIC researchers are working to understand how neutralizing antibodies coordinate with the immune system’s response.
Some people feel that the worst days of the COVID-19 pandemic are long gone, but Americans continue to die from this epidemic, and many people around the world will not receive the COVID-19 vaccine for several years.
Schendel said, “We have so many people in the United States and around the world who are not vaccinated. If we can design better monoclonal antibody therapies, there will be therapies available to them.”
Reference: Researchers combine different human antibodies to target SARS-CoV-2.
Kathryn M. Hastie et al. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: A global consortium study. Science, 2021, doi:10.1126/science.abh2315.
Researchers combine different human antibodies to target SARS-CoV-2.
(source:internet, reference only)
Disclaimer of medicaltrend.org
Important Note: The information provided is for informational purposes only and should not be considered as medical advice.