COVID-19 Delta Variant Study: Resistance to neutralizing antibodies
- Aspirin: Study Finds Greater Benefits for These Colorectal Cancer Patients
- Cancer Can Occur Without Genetic Mutations?
- Statins Lower Blood Lipids: How Long is a Course?
- Warning: Smartwatch Blood Sugar Measurement Deemed Dangerous
- Mifepristone: A Safe and Effective Abortion Option Amidst Controversy
- Asbestos Detected in Buildings Damaged in Ukraine: Analyzed by Japanese Company
COVID-19 Delta Variant Study: Resistance to neutralizing antibodies
- Israel new drug for COVID-19: EXO-CD24 can reduce deaths by 50%
- COVID-19 vaccines for children under 12 will be available soon
- Breakthrough infection of Delta: No difference from regular COVID-19 cases
- French research: ADE occurred in Delta variant and many doubts on it
- The viral load of Delta variant is 1260 times the original COVID-19 strain
COVID-19 Delta Variant Study: Resistance to neutralizing antibodies. Comprehensive study of COVID-19 Delta variants: resistance to neutralizing antibodies.
During the current SARS-CoV-2 pandemic, multiple mutations have accumulated in the viral genome, and at least five variants are considered to be harmful to human society. The newly emerged VOC B.1.617.2 lineage (Delta variant) is closely related to the large-scale surge of the COVID-19 epidemic in India in the spring of 2021, and has now become the most threatening variant of the COVID-19 virus. However, its virological properties are still unclear.
On June 17, 2021, the preprint platform bioRxiv launched articles from research institutions such as the University of Tokyo, Miyazaki University, Kumamoto University, etc. titled: SARS-CoV-2 spike P681R mutation enhances and accelerates viral fusion.

The paper shows:
1. Delta variant is a virus strain that is highly fused and easy to form syncytia. The P681R mutation in the spike protein is highly conserved in this lineage.
2. Although the P681R mutation reduces the infectivity of the virus, this mutation produces resistance to neutralizing antibodies.
3. Further mechanism studies have shown that P681R mutation promotes furin-mediated cleavage of spike protein (S protein) and accelerates cell-cell fusion.
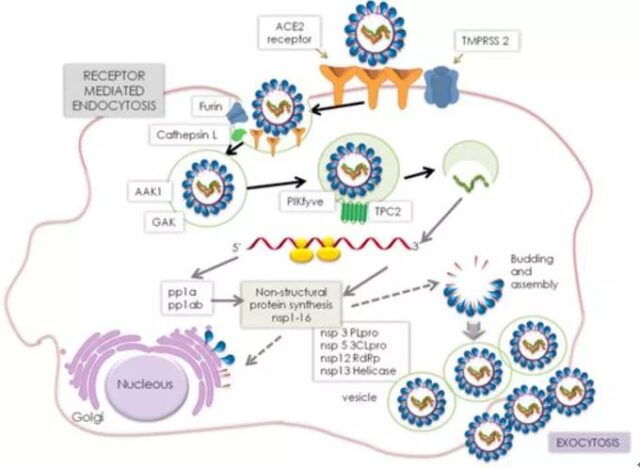
New coronavirus infection route
New coronavirus infection process:
- The first step: the binding of the viral spike glycoprotein (S protein) to the host angiotensin converting enzyme 2 (ACE2) receptor;
- The second step, host transmembrane serine protease 2 (TMPRSS2) cleavage and fusion of viral spike glycoprotein;
- In the third step, the virus fuses into the host cell membrane.
Delta variants can significantly cause syncytium formation
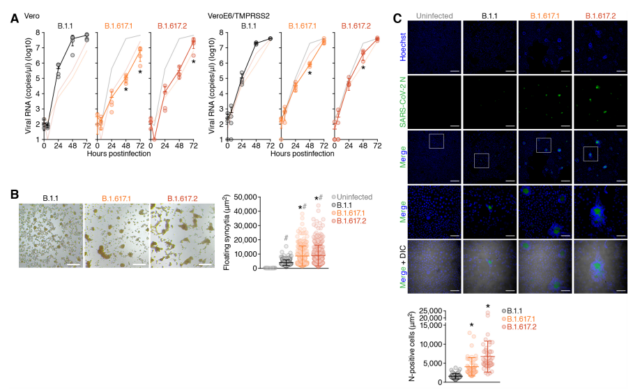
By comparing the mutant virus strains B.1.617.1, B.1.617.2 (Delta variant) and the B.1.1 isolate carrying D614G, the researchers found that in Vero cells, B.1.617.1 and B.1.617.2 The growth of the variant was significantly lower than that of the B.1.1 isolate. In particular, the viral RNA level of the B.1.617 variant was more than 150 times lower than that of the B.1.1 isolate at 48 hours after infection.
Microscopic observation showed that the VeroE6/TMPRSS2 cultures infected with these three viruses all formed syncytia. It is worth noting that, compared with the B.1.1 virus, the two B.1.617 viruses formed larger syncytia. By measuring the size of floating syncytia in cultures infected with VeroE6/TMPRSS2, the syncytia formed by B.1.617.1 and B.1.617.2 (Delta variants) infection was significantly (2.3 times and 2.7 times) larger than that of B.1.1 infection. Syncytia.
These results indicate that compared with the B.1.1 virus carrying D614G, the B.1.617 lineage (Delta variant) is more likely to form syncytia and prefer cell-cell infection.
P681R mutation is the determinant of promoting S protein-mediated fusion
The P681R mutation is a unique feature of the B.1.617 lineage (Delta variant), which is located near the FCS (furin cleavage site) of the SARS-CoV-2 S protein. The researchers produced an artificial virus carrying P681R through reverse genetics and conducted virological experiments.
Western blot of the prepared pseudovirus showed that the P681R mutation significantly increased the level of cleaved S2 subunits. A single round of pseudovirus infection test was performed with target HOS cells with or without TMPRSS2 expression.
Without TMPRSS2, the infectivity of the pseudovirus carrying P681R/D614G is equivalent to that of the D614G pseudovirus. In the presence of TMPRSS2, compared with TMPRSS2 empty target cells, the infectivity of D614G pseudovirus is increased by 5.0-6.3 times. These data indicate that the P681R mutation attenuates the infectivity of cell-free viruses in the presence of TMPRSS2.
Cell-based fusion experiments were used to study the effect of P681R mutation on virus fusion. In effector cells (i.e. S protein-expressing cells), although D614G/P681R S protein expression level was comparable to D614G S, the level of fragmented S2 subunit of D614G/P681R mutant was significantly higher than that of D614G S2 subunit.
Cell-based fusion analysis using target cells without TMPRSS2 showed that the fusion origin of D614G/P681RS was 2.1 times higher than that of D614G S, and the initial fusion speed was 2.8 times faster than D614G S. These data indicate that the P681R mutation enhances and accelerates SARS-CoV-2 S-mediated fusion.
P681R mutation is resistant to NAbs-mediated antiviral immunity
The results of the neutralization test showed that the D614G/P681R artificial virus has partial (1.2-1.5 times) resistance to the three monoclonal antibodies against the SARS-CoV-2 S protein RBD. In addition, a neutralization test with 19 BNT162b2 vaccine sera showed that D614G/P681R artificial virus has significant resistance to vaccine-induced NAbs.
Contrary to the neutralizing activity against cell-free viruses, SARS-CoV-2 S fusion experiments (artificial virus and effector cells (S-expressing cells) treated with NAbs or serum) showed that cell-cell infections have strong resistance to NAbs And the insensitivity of NAbs to cell-cell infection does not depend on the P681R mutation.
Taken together, these findings indicate that the P681R mutation confers NAbs resistance to cell-free virus particles, and cell-cell infections are resistant to NAbs-mediated antiviral effects compared to cell-free infections.
Since the outbreak of the new coronavirus in December 2019, a variety of mutant virus strains have emerged. The mutant virus strain B.1.617 (Delta variant) that appeared in India in the spring of 2021 has attracted widespread attention because of its neutralizing antibody resistance.
In this paper, through the study of the B.1.617 variant, the highly conserved P681R mutation in this pedigree was determined. Through in-depth research, it is found that the P681R mutation promotes furin-mediated cleavage of the spike protein, accelerates cell-cell fusion, and promotes B.1.617 to develop resistance to neutralizing antibodies.
(source:internet, reference only)
Disclaimer of medicaltrend.org
Important Note: The information provided is for informational purposes only and should not be considered as medical advice.