RNA-protein binding site prediction
- Normal Liver Cells Found to Promote Cancer Metastasis to the Liver
- Nearly 80% Complete Remission: Breakthrough in ADC Anti-Tumor Treatment
- Vaccination Against Common Diseases May Prevent Dementia!
- New Alzheimer’s Disease (AD) Diagnosis and Staging Criteria
- Breakthrough in Alzheimer’s Disease: New Nasal Spray Halts Cognitive Decline by Targeting Toxic Protein
- Can the Tap Water at the Paris Olympics be Drunk Directly?
RNA-protein binding site prediction
RNA-protein binding site prediction. RNA binding proteins (RBPs) play a vital role in various biological processes, and their binding sites on RNA can provide insights into the mechanisms behind diseases involving RBPs. Therefore, how to identify the RBP binding site on RNA is very important for subsequent analysis. Today, the editor will introduce to you a deep learning-based RNA-protein binding site prediction website: RBPsuite, which is an easy-to-use web server for predicting RBP binding sites on linear and circular RNAs.
RBPsuite first divides the input RNA sequence into 101 nucleotide fragments and scores the interaction between the fragments and RBP. RBPsuite further detects the verified motifs on the binding fragments to give a distribution of binding scores along the full-length sequence. For linear RNAs, RBPsuite uses iDeepS to predict their binding scores with RBP; for circular RNAs (circRNA), RBPsuite uses CRIP to predict their binding scores with RBP.
Ado,
On the link: http://www.csbio.sjtu.edu.cn/bioinf/RBPsuite/, open the main page according to the official website address as follows.
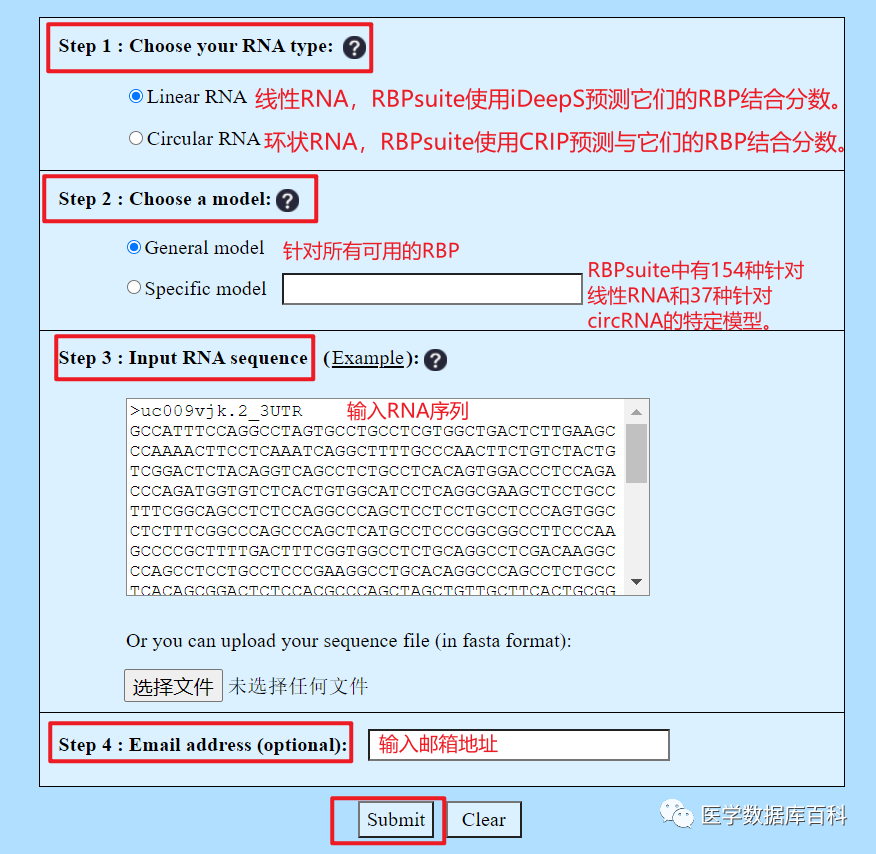
RBPsuite requires three types of input: RNA type, prediction model and RNA sequence. The email address is optional. If an email is provided, a notification will be sent to that email when the job is completed.
Step 1. Select RNA type
Linear RNA and circRNA are predicted by different models. If you are not sure which RNA type the input sequence belongs to, you can first use WebCircRNA (https://rth.dk/resources/webcircrna/submit) to evaluate the potential of circRNA, and then select the RNA type.
Step 2. Choose a prediction model
RBPsuite provides two types of predictive models: conventional models (all available RBPs) and specific models (a specific RBP). If the protein name is known exactly, a specific model is used. Otherwise, a conventional model is used, which will predict the interaction scores between all RBPs and the input RNA.
Step 3. Input RNA sequence
We can input RNA sequence or upload sequence file in FASTA format.
Step 4. Email (optional)
This step is optional. But it is recommended to enter the email address, RBPsuite will send the result to the email. This way we can save the result without losing it.
Let’s take linear RNA and conventional models as an example to illustrate the brief use of this database.
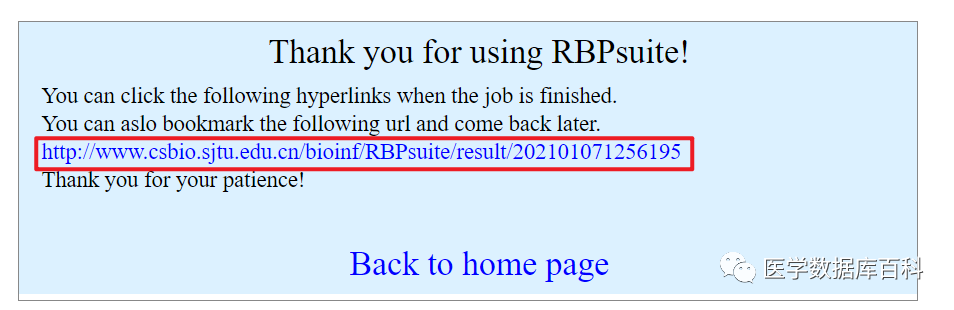
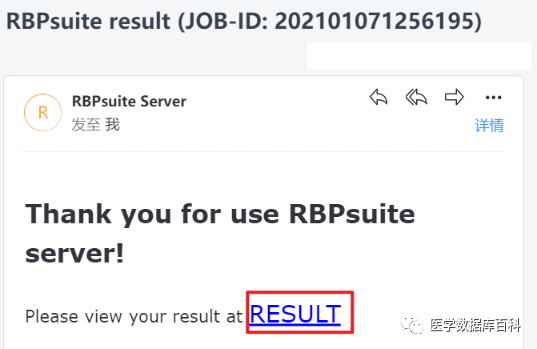
After submission, jump to the result page, we can click this URL or the RESULT in the email below to view the result.
The result will show the interaction score between the input RNA and all RBPs. For a single RBP, we can click on the RBP of interest to view more detailed information about the prediction result. Let’s take RBP: CSTF2T as an example to briefly explain the results.

Click CSTF2T, the following table will show the specific scores of the different segments of the input RNA sequence combined with RBP.
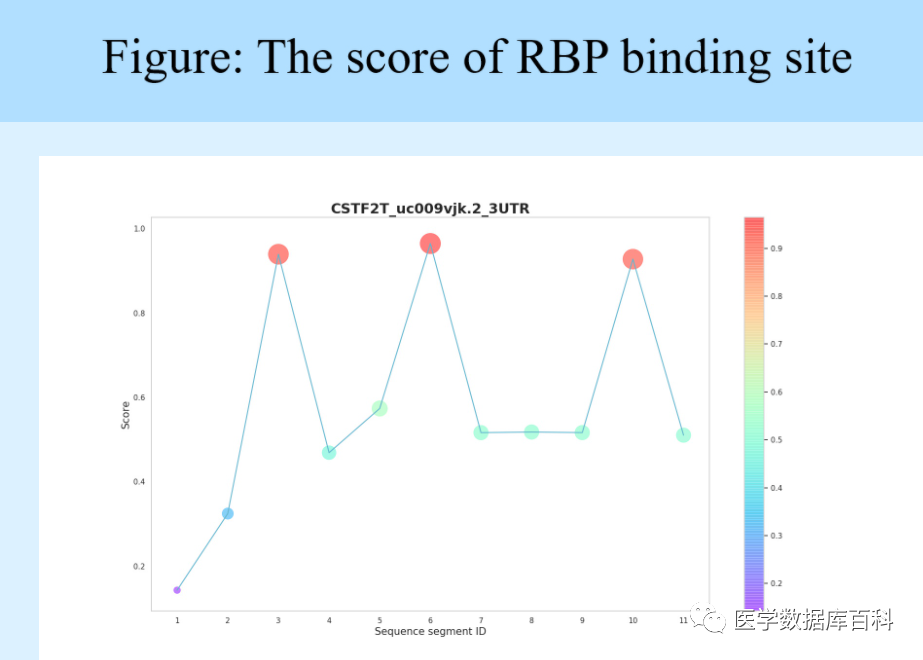
Note: In order to improve the computational efficiency, RBPsuite divides the input sequence into a variable of length 101
Overlap the segments and display the score of each segment with the RBP of interest in the table.
Of course, these results are all available for download.
(sourceinternet, reference only)
Disclaimer of medicaltrend.org



