World’s 1st electron microscopic structure of a human-derived secondary spliceosome
- Normal Liver Cells Found to Promote Cancer Metastasis to the Liver
- Nearly 80% Complete Remission: Breakthrough in ADC Anti-Tumor Treatment
- Vaccination Against Common Diseases May Prevent Dementia!
- New Alzheimer’s Disease (AD) Diagnosis and Staging Criteria
- Breakthrough in Alzheimer’s Disease: New Nasal Spray Halts Cognitive Decline by Targeting Toxic Protein
- Can the Tap Water at the Paris Olympics be Drunk Directly?
World’s 1st electron microscopic structure of a human-derived secondary spliceosome
World’s 1st electron microscopic structure of a human-derived secondary spliceosome. A major breakthrough! The research group of Yigong Shi of West Lake University published an article in “Science ” to report the electron microscopic structure of the world’s first human-derived secondary spliceosome.
On January 29, 2021, the research group of Professor Yigong Shi of West Lake University published a research paper titled “Structure of the Activated Human Minor Spliceosome” in Science (Science) , Is another major breakthrough in the study of spliceosome structure and mechanism.
In this paper, they reported for the first time the high-resolution three-dimensional structure of the minor spliceosome, which has been poorly understood in the overall study so far, and demonstrated a key conformation in the splicing reaction-the activated minor spliceosome (activated minor spliceosome). , Defined as “secondary Bact complex”), the overall resolution is up to 2.9 angstroms.
This structure shows for the first time the composition of a human-derived minor spliceosome and the recognition mechanism for rare introns (U12-dependent introns), and for the first time reveals the catalytic center and active site of the minor spliceosome, and Through structural analysis, a series of important scientific issues have been identified, including new protein components of the minor spliceosome and revealed their important role in the splicing of minor spliceosome and rare introns.
This major breakthrough enabled Shi Yigong’s research group to analyze the world’s first spliceosome structure for the first time in 2015 and the first human-sourced spliceosome structure in 2017, and once again became the world’s first to analyze secondary splicing. A team of high-resolution three-dimensional structures.

Figure 1 The latest “Science” by Shi Yigong’s research group
In higher organisms, segments of genetic code are buried in nucleic acid molecules. Among them, “invalid” genetic information has no translation function and is called an intron, while the effective genetic information that can be translated by the ribosome is called an exon. The process of “scissing” out the “invalid” introns in these genetic codes and “splicing” the “valid” exons together is called RNA splicing, which is the core step in the interpretation of the genetic code by living organisms.
The complex and sophisticated molecular machine in the nucleus-the spliceosome, is responsible for performing this step. The splicing of each cell for each gene messenger before splicing-precursor messenger RNA is very precise in time and space. Who to cut, how long to cut, when to cut, and in what order to splice the exons, each “operation” is a key issue that may change the fate of cells.
One step wrong, the results are very different, the gene expression and transmission will also be wrong, which leads to the emergence of certain congenital genetic diseases and acquired incurable diseases. Studies have found that about 35% of human genetic diseases are caused by splicing abnormalities.
From the first discovery of RNA splicing in 1977 to the beginning of this century, scientists used immunoprecipitation, gene knockout, cross-linking mass spectrometry, and the establishment of an in vitro splicing reaction system to initially establish the process of splicing assembly, activation and depolymerization. , And complex RNA splicing regulatory networks such as the interaction between protein and protein, protein and nucleic acid, and mutual regulation.
In the 1990s, scientists discovered a class of non-classical intron splicing sequences (5′ splice site is AT, 3’splice site is AC) in the research and analysis of intron sequences of higher eukaryotes. , A new spliceosome began to gradually enter the field of vision of scientists. In subsequent studies, scientists gradually determined the nucleic acid composition of this new spliceosome. Four new snRNAs, U11, U12, U4atac, and U6atac, are involved in the recognition and splicing of this type of intron.
Since this non-classical intron accounts for less than one percent, this type of intron is called a rare intron. Because of its special splicing site and the special snRNA component that recognizes this site, the The class of introns is also named AT-AC intron (AT-AC intron) or U12-type intron (U12-type intron). This new spliceosome that catalyzes the splicing of this type of intron is named Minor spliceosome (minor spliceosome).
Although the content of rare introns spliced by minor spliceosomes is extremely low, these genes are closely related to many important basic cell life activities, such as DNA replication and repair, RNA transcription and translation, and so on. However, the research on U12-dependent introns and secondary spliceosome has been developing slowly, and key scientific questions such as the protein composition of the secondary spliceosome and the regulation of remodeling have not been answered. One of the very important steps-how to capture and purify the minor spliceosome is a huge problem in the field.
Yigong Shi’s research group has been committed to the study of the three-dimensional structure of the spliceosome and the molecular mechanism of RNA splicing. Since the first high-resolution three-dimensional structure of the spliceosome was reported in 2015, it has successively analyzed the main spliceosome of Saccharomyces cerevisiae and human origin. (Major spliceosome) All basic conformations that have been identified.
These resolved spliceosomes cover the entire RNA splicing cycle, revealing the working mechanism of the spliceosome catalyzing the two-step reaction of RNA splicing at the molecular level, and provide a structural basis for understanding the processes of spliceosome assembly, activation, and depolymerization. The complete connection of the spliceosome-mediated RNA splicing process provides the clearest and most comprehensive structural information for understanding the molecular mechanism of RNA splicing (Figure 2).
At the same time, Shi Yigong’s research group also focused on the secondary spliceosome research area where research is even scarcer.
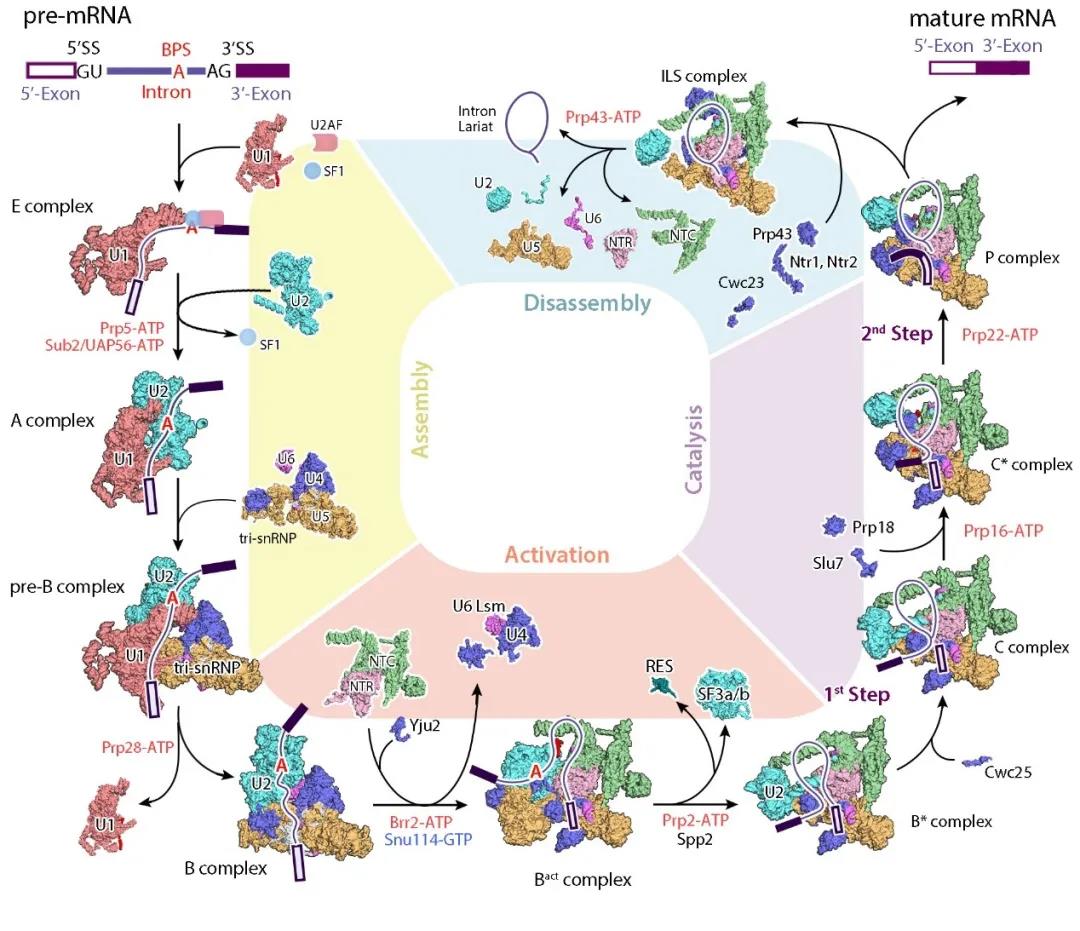
Figure 2 Summary of spliceosome structure analyzed by Shi Yigong’s research group
(Image source: Wan et.al, 2020, Annu Rev Biochem)
In the absence of any relevant literature on the capture and purification of secondary spliceosomes, it is urgent to explore and establish methods for the capture and purification of secondary spliceosomes. Dr. Bai Rui and Dr. Wan Ruixue of Shi Yigong’s research group have long been engaged in the purification and structural research of spliceosome, and have accumulated a lot of experience in spliceosome structure research.
On the basis of this research experience, combined with the specificity of the secondary spliceosome recognition site, the research group designed a new U12-dependent pre-mRNA for the first time. By improving the predecessors’ in vitro splicing reaction experimental conditions, it was successfully determined that the U12-dependent pre-mRNA has extremely high specificity and splicing efficiency. Due to the extremely low content of the secondary spliceosome, how to obtain further samples of the secondary spliceosome with good properties has become the second major difficulty in this paper.
Bai Rui and Wan Ruixue further explored and improved on the basis of the original spliceosome purification, and finally established a complete set of secondary spliceosome capture and purification methods for the first time, and successfully obtained the activated secondary spliceosome Protein samples were then used single-particle cryo-electron microscopy technology to reconstruct the world’s first cryo-electron microscopy structure of the secondary spliceosome, with an overall resolution of 2.9 angstroms, and built the first atomic model of the secondary spliceosome, including 4 RNAs and 45 proteins were obtained (Figure 3).
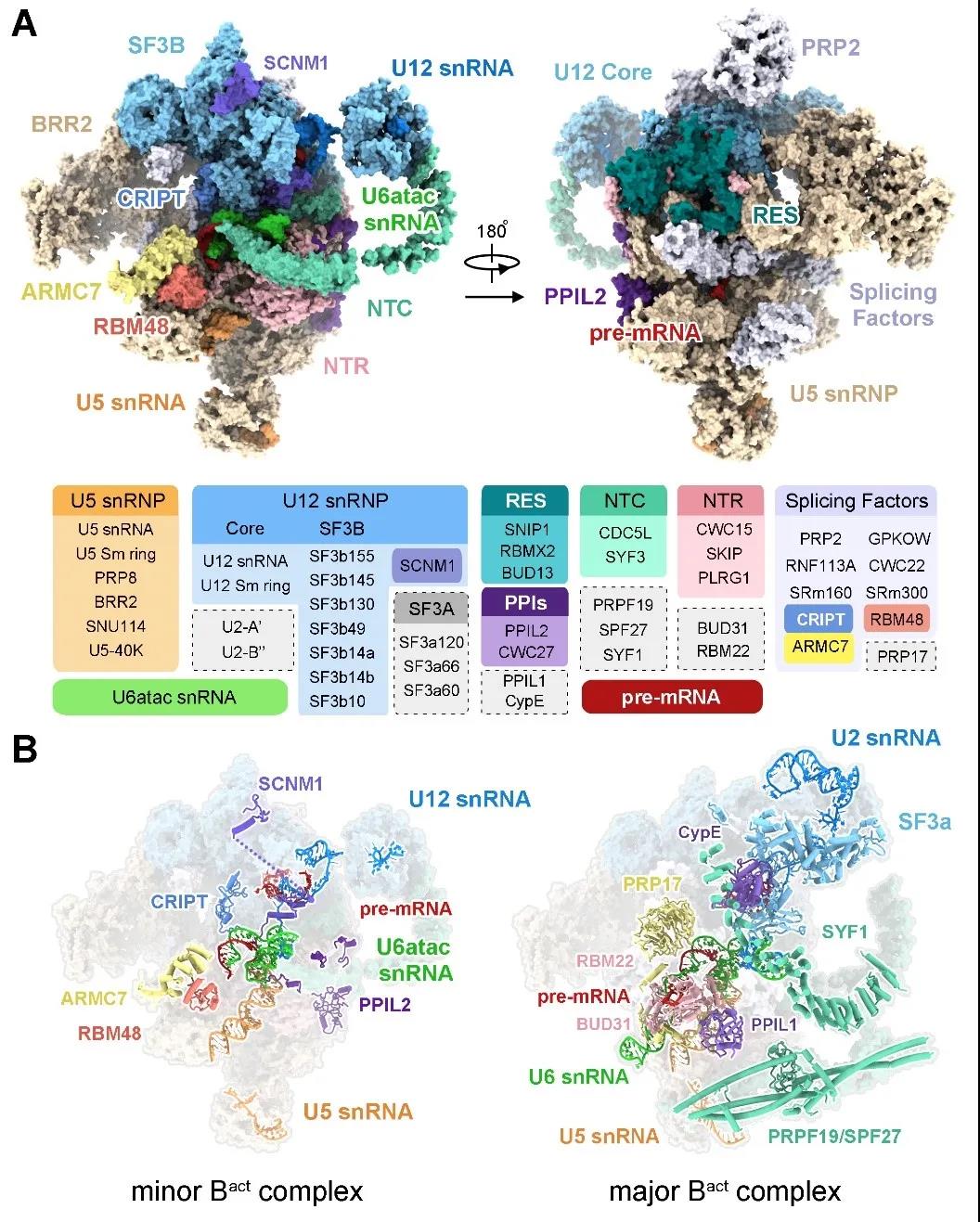
Figure 3 The three-dimensional structure of the human secondary spliceosome
The structure of the human-derived minor spliceosome Bact complex analyzed in this paper was the first to observe the three-dimensional structure of the U6atac snRNA and U12 snRNA, which are unique components of the minor spliceosome, and reveal the structural basis for recognizing the splice site of U12-type introns. The three-dimensional structure of the active site and catalytic center of the minor spliceosome is shown once. In the active site of the secondary spliceosome, two key catalytic magnesium ions are coordinated and bound by the surrounding RNA, which provides the basis for the splicing reaction. The successful analysis of this structure identified five brand-new protein components for the first time. They participate in the formation of secondary spliceosome and therefore play an important role in the splicing of U12-dependent introns.
SCNM1 and CRIPT are the unique components of U12 snRNP, which are involved in recognizing and stabilizing the splicing site on introns; RBM48 forms a complex with ARMC7 and binds to the 5’end of U6atac snRNA, binding and protecting the unique γ-formaldehyde of U6atac snRNA The base phosphate cap structure stabilizes the conformation of U6atac snRNA.
In addition, the structure of the PPIL2 protein involved in the ubiquitination pathway in the spliceosome was analyzed for the first time. The protein penetrates into the active site of the secondary spliceosome, interacts directly with U5 snRNA, and participates in stabilizing U5 snRNP and the secondary spliceosome. Catalytic center. It is worth mentioning that the “power-driven” protein Prp2 and its activator Spp2, which play a key role in the activation of the primary spliceosome, are also incorporated into the secondary spliceosome in a similar way. The first supporting evidence for a role in the spliceosome.
It is not difficult to infer from this that several ATPases that promote the structural remodeling of the main spliceosome are also involved in the transformation and remodeling between the different conformations of the secondary spliceosome, that is, the secondary spliceosome shares the structural remodeling with the main spliceosome. Protein factor (Figure 4). This article is the first to reveal the structural study of human-derived minor spliceosomes. The methods established in the paper to capture and purify minor spliceosomes, as well as the new proteins that participate in the composition of minor spliceosomes, will all be dependent on U12 The study of the molecular mechanism of RNA splicing has an important impact.
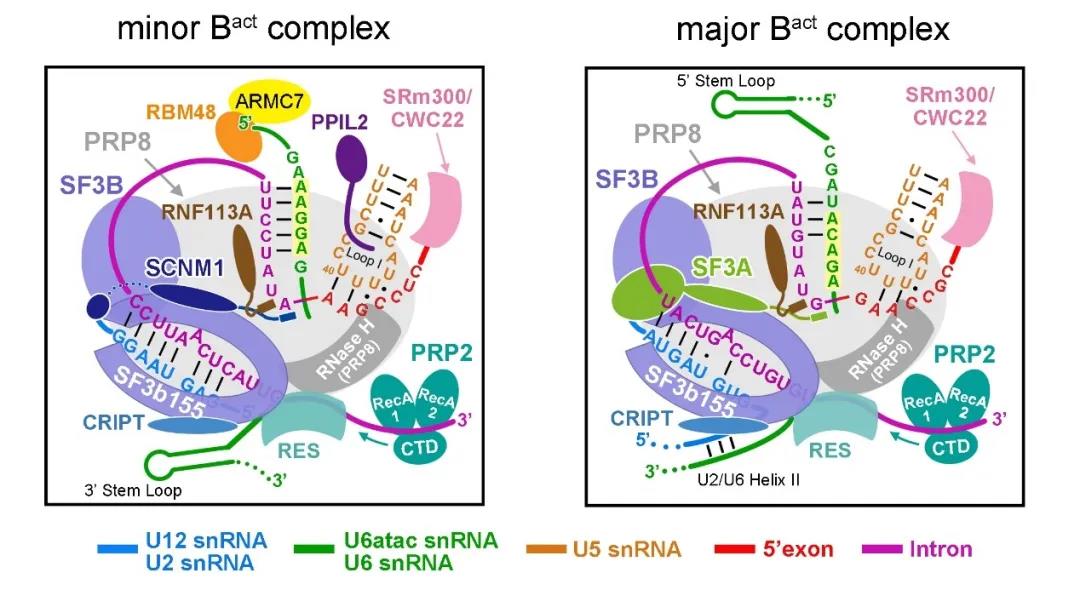
Figure 4 The structural difference between the minor spliceosome and the major spliceosome
Professor Shi Yigong, School of Life Sciences, West Lake University, and Dr. Wan Ruixue, a West Lake scholar from West Lake University’s School of Life Sciences, are the co-corresponding authors; Bai Rui, a postdoctoral fellow from West Lake University’s School of Life Sciences, and West Lake scholar Wan Ruixue are the co-first authors of this article; Wang Lin, a postdoctoral fellow in the School of Life Sciences, Tsinghua University, participated in some biochemical research; Xu Kui, a doctoral student in the School of Life Sciences, Tsinghua University, and Zhang Qiangfeng, a researcher from the School of Life Sciences, Tsinghua University, provided assistance in the construction of the structure model; Dr. Lei Jianlin, director of the cryo-EM platform at Tsinghua University The electron microscope data collection provided assistance.
The electron microscopy data was collected on the cryo-electron microscopy platform of Tsinghua University, and the calculation was supported by the high-performance computing platform of Tsinghua University and the National Protein Facilities Experimental Technology Center (Beijing). The results of this research came from the West Lake Laboratory and received funding support from the West Lake Education Foundation, Beijing Advanced Innovation Center for Structural Biology (Tsinghua) and the National Natural Science Foundation of China.
~~ World’s 1st electron microscopic structure of a human-derived secondary spliceosome ~~
(source:internet, reference only)
Disclaimer of medicaltrend.org



