Glioma: Necessary to detect the copy number variation of chromosome 7/10
- A Single US$2.15-Million Injection to Block 90% of Cancer Cell Formation
- WIV: Prevention of New Disease X and Investigation of the Origin of COVID-19
- Why Botulinum Toxin Reigns as One of the Deadliest Poisons?
- FDA Approves Pfizer’s One-Time Gene Therapy for Hemophilia B: $3.5 Million per Dose
- Aspirin: Study Finds Greater Benefits for These Colorectal Cancer Patients
- Cancer Can Occur Without Genetic Mutations?
Glioma: Necessary to detect the copy number variation of chromosome 7/10
Glioma: Necessary to detect the copy number variation of chromosome 7/10, Recommendations in cIMPACT updates 3 and 6: In adult IDH wild-type diffuse astroglioma, if there is one of the following three genetic parameters (TERT promoter mutation, EGFR amplification, accompanied by the acquisition of the entire chromosome 7 And the loss of the entire chromosome 10) can be located at WHO level IV.
Summary
Recommendations in cIMPACT updates 3 and 6: In adult IDH wild-type diffuse astroglioma, if there is one of the following three genetic parameters (TERT promoter mutation, EGFR amplification, accompanied by the acquisition of the entire chromosome 7 And the loss of the entire chromosome 10) can be located at WHO level IV. Therefore, it is necessary for patients with glioma to detect the copy number variation of chromosome 7/10. The technique commonly used in clinic to detect the acquisition of the entire chromosome 7 and the loss of the entire chromosome 10 is FISH.
What is chromosome copy number variation?
DNA copy number variation (CNV) is an important part of genetic variation. Compared with single nucleotide polymorphism (SNP), it has a greater impact on the genome. The role of germline (germ cell) CNV characteristics in a variety of genetic diseases has been widely understood, however, there are relatively few studies on somatic CNV.
Somatic chromosome copy number changes (SCNA), including the gains and losses of the entire chromosome or the entire arm, affect a large area of the genome that may contain known oncogenes and tumor suppressor genes. Such genomic instability events promote the occurrence and development of tumors by causing the rapid accumulation of other possible driver gene mutations [1].
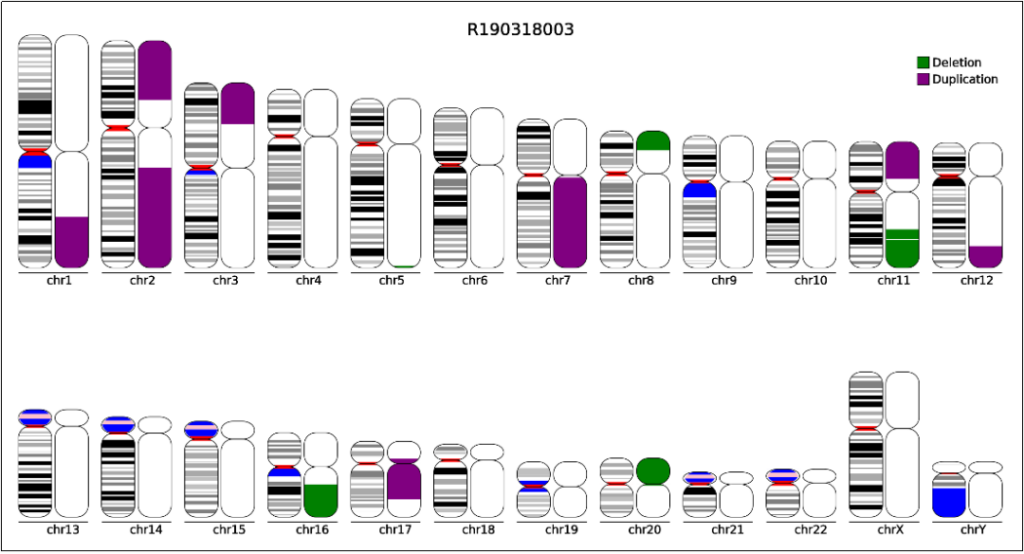
Figure 1. Schematic diagram of genome chromosome copy number variation
The copy number variation of chromosome 7/10, a molecular marker of glioma
Glioma is the most common intracranial primary malignant tumor. According to the 2016 WHO classification of central nervous system tumors, the final diagnosis of glioma requires molecular pathology information. Common low-grade glioma types include diffuse astrocytoma IDH wild type, diffuse astrocytoma IDH mutant ATRX deletion and TP53 mutant type, oligodendroglioma IDH mutation and 1p/19q co-deletion type [2].
With the development of high-throughput technology, it has been found that most of the IDH wild-type low-grade gliomas have genome chromosome copy number variation similar to those in the glioblastoma IDH wild-type. It is worth emphasizing that more than 50% of IDH wild-type low-grade gliomas have both the amplification of chromosome 7 and the deletion of chromosome 10, while the copy number of chromosome 7/10 in patients with IDH wild-type glioblastoma The proportion is higher (up to 80%).
Studies have reported that patients with chromosomal imbalance such as 7p or 7q increase and 10p or 10q loss have a lower survival rate for patients with WHO grade II or III IDH wild-type astrocytoma. However, most studies reported that the overall increase of chromosome 7 and the overall loss of chromosome 10 (+7/-10) are the main characteristics. The +7/-10 combination feature has poor clinical prognosis in IDH wild-type astrocytoma patients [3].
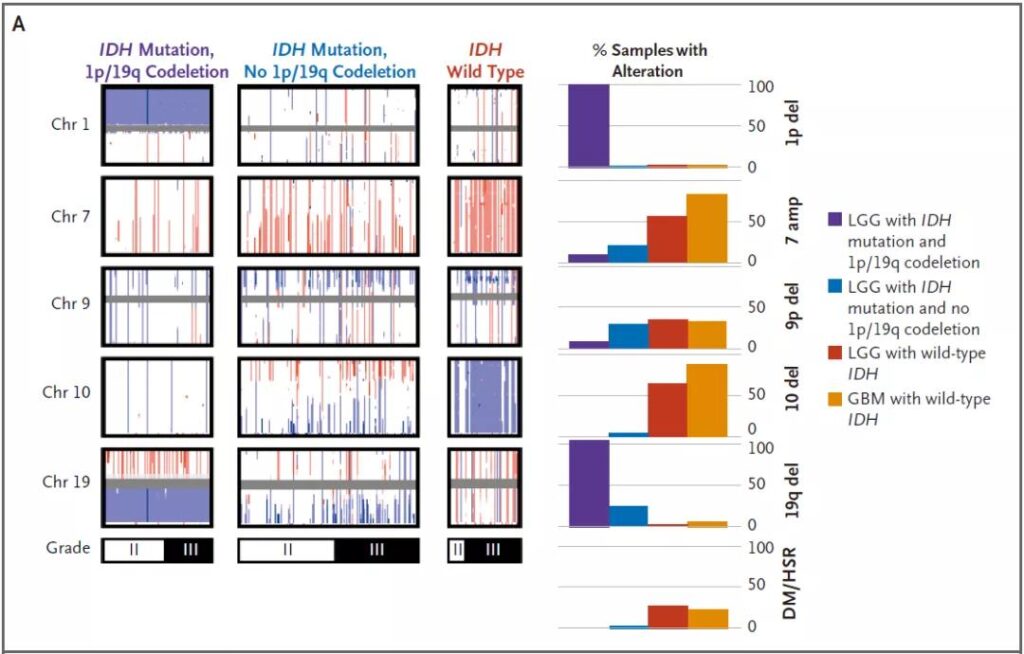
Figure 2. Chromosome copy number variation in glioma
Clinical significance of detecting chromosome 7/10 copy number variation in glioma
Researchers found that the prognosis and clinical behavior of most IDH wild-type low-grade gliomas are comparable to those of primary glioblastoma, because these low-grade gliomas are similar to those of primary glioblastoma. The same molecular characteristics, such as 7/10 chromosome copy number variation and TERT promoter mutation [4].
Recommendations in cIMPACT updates 3 and 6: In adult IDH wild-type diffuse astroglioma, if there is one of the following three genetic parameters (TERT promoter mutation, EGFR amplification, accompanied by the acquisition of the entire chromosome 7 And the loss of the entire chromosome 10) can be located at WHO level IV. At the same time, it is recommended to name it “diffuse astrocytic glioma, IDH wild-type, with molecular characteristics of glioblastoma, WHO grade IV” [5].
This shows that IDH wild-type low-grade glioma is a high-grade glioma that is underestimated by histological typing. cIMPACT updates 3 and 6 recommend the use of three genetic parameters as the criteria for diagnosing IDH wild-type glioblastoma, and the naming can be simplified for effective inclusion in relevant clinical trials.
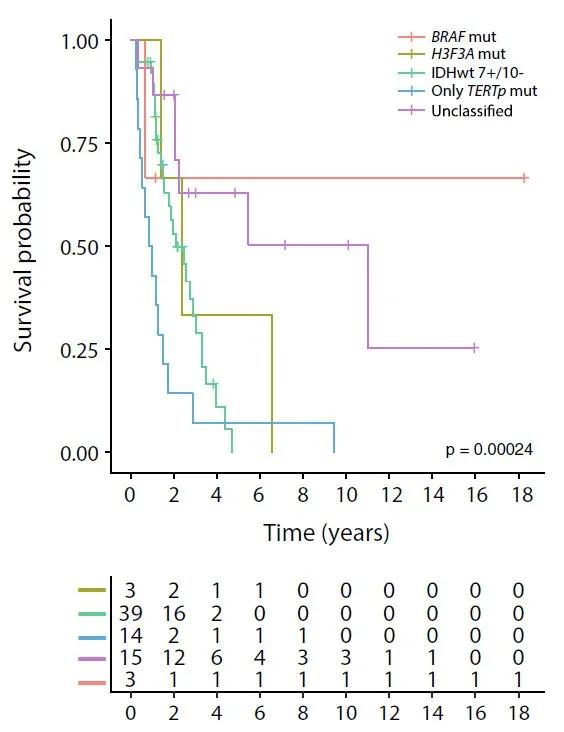
Figure 3. Overall survival rate of 74 patients stratified by molecular subgroup
What technology can detect chromosome copy number variation?
At present, CNV detection technologies include fluorescence in situ hybridization (FISH), single nucleotide polymorphism gene chip (SNP array) and fast-developing low-depth whole-genome sequencing technologies. These technologies have their own advantages and disadvantages (Table 1 ).
With the help of next-generation sequencing technology and corresponding experimental strategies, CNV can be discovered and accurately located. Although there are a variety of techniques for detecting chromosome copy number variation, the most common technique for detecting chromosome copy number variation in glioma 7/10 is FISH.
(source:internet, reference only)
Disclaimer of medicaltrend.org