RNA virus polymerase has a characteristic region related to host adaptation
- Normal Liver Cells Found to Promote Cancer Metastasis to the Liver
- Nearly 80% Complete Remission: Breakthrough in ADC Anti-Tumor Treatment
- Vaccination Against Common Diseases May Prevent Dementia!
- New Alzheimer’s Disease (AD) Diagnosis and Staging Criteria
- Breakthrough in Alzheimer’s Disease: New Nasal Spray Halts Cognitive Decline by Targeting Toxic Protein
- Can the Tap Water at the Paris Olympics be Drunk Directly?
RNA virus polymerase has a characteristic region related to host adaptation
RNA virus polymerase has a characteristic region related to host adaptation. RNA-dependent RNA polymerase (RNA-dependent RNA polymerase, referred to as RdRP) encoded by RNA viruses is a unique type of nucleic acid polymerase that plays a central role in viral genome replication and transcription, and is a hot target for antiviral drug research.
Virus RdRP can be fused with other functional domains or co-folded with other viral proteins, and its overall structural diversity is high, but the three-dimensional structure of its catalytic core region is relatively conservative, so RdRP has both diversity and conservation.
Since the host range of RNA viruses includes almost all cell forms of life, in the process of co-evolution with different hosts, RdRP is the most conserved protein of RNA viruses, and the relationship between it and the adaptability of the virus to the host is not clear.
Flaviviruses include Japanese encephalitis virus (JEV), dengue virus (DENV), Zika virus, tick-borne encephalitis virus (TBEV), etc. Human pathogens are a kind of single-stranded positive-stranded RNA viruses with a wide distribution and a wide variety of species. They belong to the flaviviridae together with hepatitis C virus and classical swine fever virus.
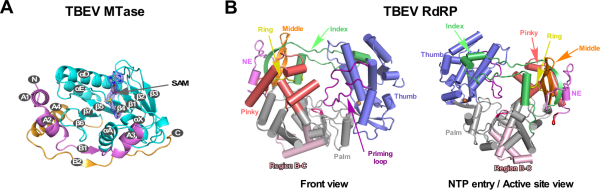
Flaviviruses are mostly spread by blood-sucking arthropods such as mosquitoes and ticks, which can cause human encephalitis or hemorrhagic diseases and pose a major threat to human health. The RdRP of flavivirus is located at the carboxyl end of the virus-encoded non-structural protein NS5 and forms a natural fusion with the amino-terminal methyltransferase (MTase). Previously, there have been several reports on the three-dimensional structure of NS5 of mosquito-borne flavivirus, including the structure of the full-length protein, MTase region and RdRP region, while the full-length structure of the NS5 protein of tick-borne flavivirus and the three-dimensional structure of RdRP have not been resolved.
Gong Peng’s team, a researcher at the Wuhan Institute of Virology, Chinese Academy of Sciences, has long been engaged in the study of the catalysis and regulation mechanism of viral RdRP. Previously, they have analyzed the crystal structures of the full-length NS5 protein of JEV and DENV (Lu and Gong, PLoS Pathogens 2013; Wu et al., PLoS Pathogens 2020), and in collaboration with the team of Wuhan Institute of Virology researcher Zhang Bo, systematically revealed the conformational diversity and conservation of NS5 and the molecular mechanism of MTase regulating RdRP (Li et al. PLoS Neglected Tropical Diseases 2014; Wu, et al. Journal of Virology 2015; Wu et al., PLoS Pathogens 2020).
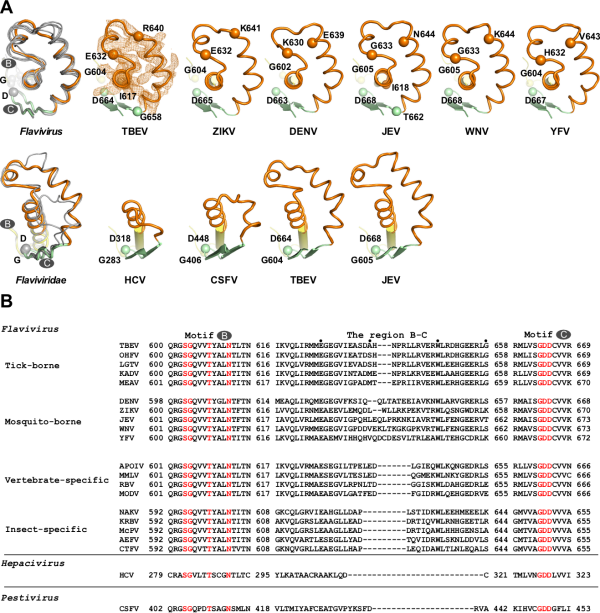
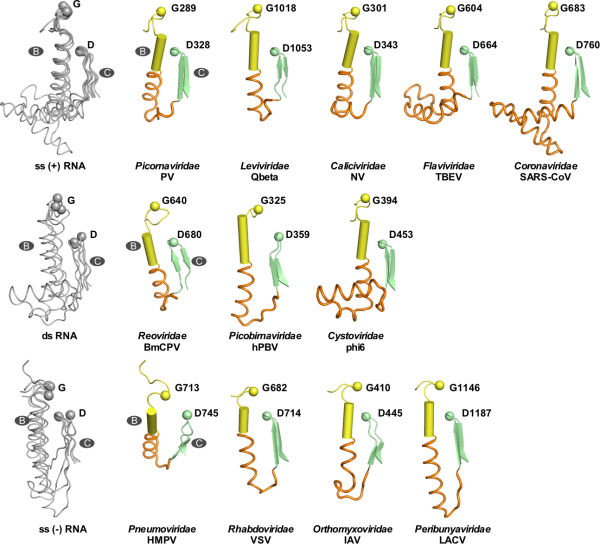
Researchers have recently analyzed the crystal structure of TBEV MTase with a resolution of 1.9 angstroms (PDB number 7D6M, Figure 1A) and the crystal structure of TBEV RdRP with a resolution of 3.2 angstroms (PDB number 7D6N, Figure 1B), and obtained the first tick transmission. Three-dimensional structure information of the flavivirus RdRP. Through sequence analysis of the flavivirus RdRP and comparison with the structure of the known mosquito-borne flavivirus RdRP, it is found that there is a region of concern between the conservative catalytic motifs (motif) B and C of RdRP (hereinafter referred to as BC link Area). This region is located at the bottom of the palm area of RdRP and exposed on the surface of the protein, and has obvious host-related diversity in different host classifications of the Flavivirus (Figure 1B and Figure 2).
The study designed substitution mutations in the B-C junction between TBEV and JEV viruses, and confirmed at the enzymatic level that mutations do not have an essential effect on the catalytic function of RdRP (Figure 3A, B). Gong Peng’s team worked with Wuhan Institute of Virology researchers Wang Hanzhong/Zheng Zhenhua’s team and Zhang Bo’s team to evaluate the effects of mutations on TBEV and JEV at the cellular level. The study showed that the mutated viruses in the two virus systems could not maintain cell proliferation (Figure 3C, D).
These results suggest that the B-C junction region may be involved in important processes related to virus proliferation other than RdRP catalysis. Through systematic analysis of the structure and sequence of the BC junction region of the representative RdRP in positive-strand, negative-strand and double-stranded RNA viruses, it is found that this region has a high diversity in structure and sequence length in the large family of RNA viruses (Figure 4), suggesting that the BC junction region of RdRP is likely to be a host adaptation hotspot shared by RNA viruses. This study provides important clues for the research on the regulatory mechanism of viral RdRP and the host adaptation research related to RdRP.
The research work was supported by the National Key Research and Development Program “Study on the Co-infection and Co-pathogenic Mechanism of Important Livestock and Poultry Pathogens” (the project leader is Ding Chan, a researcher at the Shanghai Veterinary Research Institute of the Chinese Academy of Agricultural Sciences) and the National Natural Science Foundation of China.
Doctoral student Yang Jieyu of Wuhan Institute of Virology and postdoctoral fellow Jing Xuping are the co-first authors of the paper. They mainly completed the structure and enzymology research. Gong Peng, Zheng Zhenhua and Zhang Bo are the co-corresponding authors of the paper. Wang Hanzhong/Zheng Zhenhua’s team is the experimenter Yi Wenfu, master student Yao Chen, and Zhang Bo’s postdoctoral fellow Li Xiaodan have completed TBEV and JEV virology research work respectively. Related research results were published online in Nucleic Acids Research (“Nucleic Acids Research”).
(source:internet, reference only)
Disclaimer of medicaltrend.org



