The diversity of tumor cells drives the reorganization of the liver cancer microenvironment
- What are the WHO recommendations for Japanese encephalitis vaccines?
- Individuals Carrying Two APOE4 Copies Likely to Develop Alzheimer’s Disease Symptoms
- What Is The Role of Lactic Acid in Tumor Growth and Therapy Resistance?
- The Enigma of Beethoven’s Deafness: Unveiling the Role of Lead Poisoning
- World First Autologous Regenerated Islet Transplantation Successful
- FDA Approved Opdualag: The First Immunotherapy Targeting LAG-3
The diversity of tumor cells drives the reorganization of the liver cancer microenvironment
- AstraZeneca Admits for the First Time that its COVID Vaccine Has Blood Clot Side Effects
- Was COVID virus leaked from the Chinese WIV lab?
- HIV Cure Research: New Study Links Viral DNA Levels to Spontaneous Control
- FDA has mandated a top-level black box warning for all marketed CAR-T therapies
- Can people with high blood pressure eat peanuts?
- What is the difference between dopamine and dobutamine?
- What is the difference between Atorvastatin and Rosuvastatin?
- How long can the patient live after heart stent surgery?
The diversity of tumor cells drives the reorganization of the liver cancer microenvironment.
The biological diversity of tumor cells is a key factor in the treatment failure of solid malignant tumors and the poor prognosis of patients.
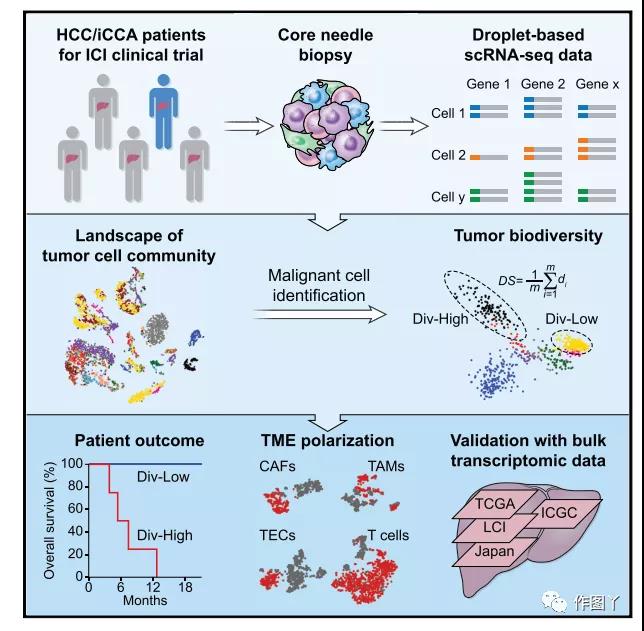
A paper titled “Tumor Cell Biodiversity Drives Microenvironmental Reprogramming in Liver Cancer” published by the National Cancer Institute team in Cancer Cell.
This work analyzed the single-cell data of 19 patients with liver cancer and found that there is a diversity of malignant cells among tumors, and there are also large differences in the tumor microenvironment, and found consistency in the independent data sets of 765 liver cancer patients. the result of.
Background introduction
During the occurrence and development of cancer, there are often complex genomics and tumor microenvironments among various tissue types.
These diverse characteristics make the development of molecular targeted therapy extremely difficult.
At the same time, it is very difficult to effectively identify driver genes among thousands of passenger genes, which also limits the predictability of precision tumor treatment strategies.
Therefore, the diversity of cancer genomes, especially intratumoral heterogeneity (ITH), will lead to the failure of tumor treatment.
The molecular characteristics of tumor cells at the single-cell level may help to identify and clarify the driving factors of cancer and the time sequence of cancer development.
main conclusion
1. HCC (hepatocellular carcinoma) and iCCA (intrahepatic cholangiocarcinoma) have varying degrees of transcriptional diversity;
2. Tumor transcription diversity is related to patient prognosis;
3. Tumor-derived vascular endothelial growth factor drives the reorganization of the microenvironment;
4. Tumor T cells from higher heterogeneity have lower cytolytic activity.
Result analysis:
01 Single-cell transcriptome data analysis of primary liver cancer
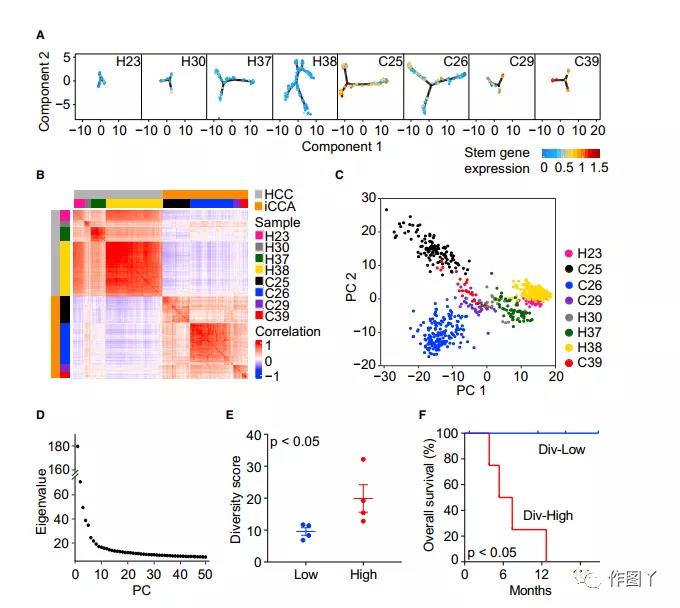
To test the diversity of tumor cells, the team performed single-cell RNA sequencing (scRNAseq) in newly isolated HCC and iCCA patients in clinical trials of immune checkpoint inhibitors. There are a total of 12 patients in the training set (set 1), and a total of 7 patients in the validation set (set 2).
This work first standardized the data. Cells with more than 500 genes detected in each cell were retained. A single-cell transcriptome of 5115 cells in set 1 was obtained, and then the 2244 genes with the largest differences among all cells were obtained. Principal component analysis.
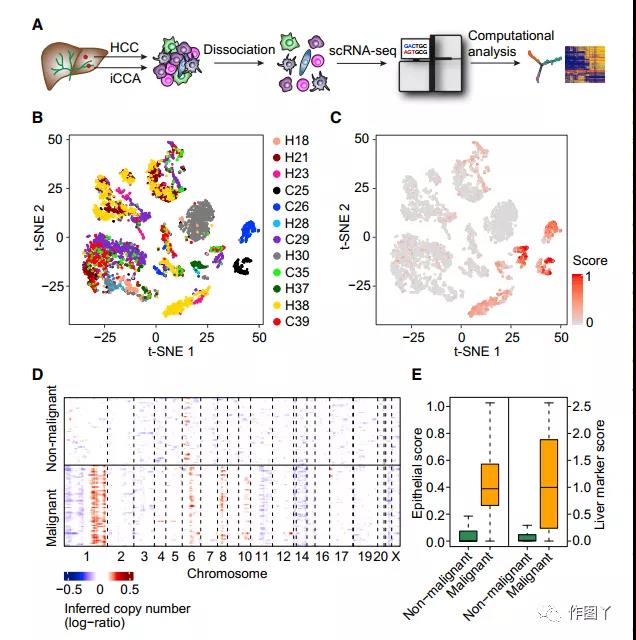
The t-SNE analysis showed that some of the patient’s cells formed different clusters, while others overlapped with each other. It was also found that some cells express high levels of epithelial marker genes, suggesting that they are potentially proto-cancerous (Figure 1C).
By inferring large-scale chromosomal copy number variations (CNVs) based on the transcriptome, we further distinguished 702 malignant cells and 4380 non-malignant cells (Figure 1D), and successfully enriched malignant and non-malignant cells based on epithelial marker genes and liver marker genes. For malignant cells, the expression of these marker genes is highly consistent with the karyotypes of malignant and non-malignant cells (Figure 1E).
02 Heterogeneity of liver cancer
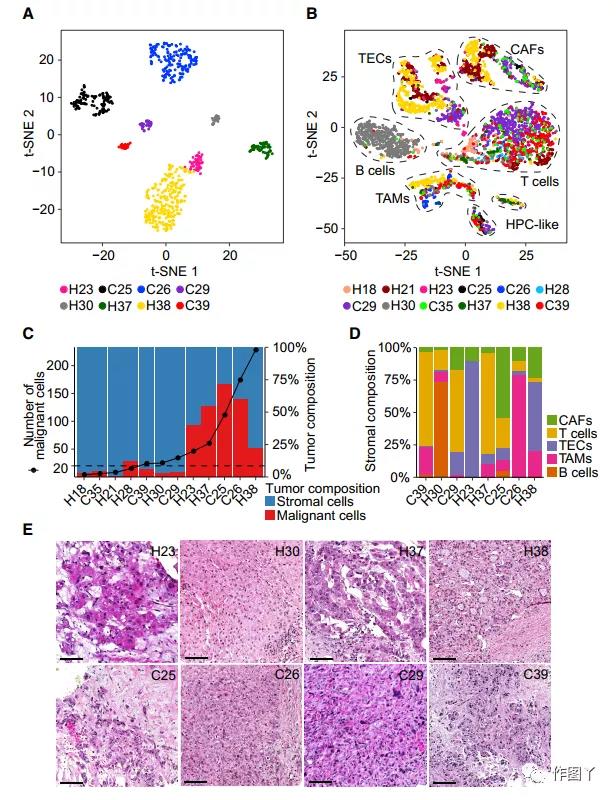
Through t-SNE analysis of malignant and non-malignant cells, it was found that malignant cells formed patient-specific cell clusters. At the same time, it was also observed that the cells inside the tumor were also heterogeneous, indicating that there is transcriptome heterogeneity between and within tumors. (Figure 2A).
In contrast, non-malignant cells are mainly grouped according to cell type (Figure 2B). The article compared the changes in the ratio of malignant and non-malignant cells in tumor patients (Figure 2C), and selected 8 samples with more than 20 malignant cells for analysis.
In these 8 patients, the composition of stromal cells was also highly heterogeneous, highlighting the complex TME in the biopsy sample (Figure 2D). Among them, H23, H30, H37, and H38 are HCC, and C25, C26, C29, and C39 are iCCA (Figure 2E).
03 Intratumoral heterogeneity (ITH) is related to prognosis
By comparing the average expression levels of 10 liver cancer stem cell-related genes EPCAM, KRT19, PROM1, ALDH1A1, CD24, ANPEP, CD44, ICAM1, CD47, and SOX9, the article determined the relationship between the proximity similarity of tumor cells and cell stem cells (Figure 3A). And through the correlation analysis of all malignant cells, the diversity of the transcriptome in the tumor was further revealed (Figure 3B).
In order to further determine the level of ITH, the article developed a method for calculating tumor cell-specific transcriptome diversity scores. Using PCs instead of the original malignant cell gene expression profile to measure the diversity of each tumor (ie ITH), to capture the main information and reduce noise (Figure 3C).
The top 30 individuals were selected according to the eigenvalue ranking test (Figure 3D). Then, according to the diversity score of the tumor samples, the 8 tumor samples were divided into a high diversity (div-high) group and a low diversity (div-low) group (Figure 3E). The overall survival and progression-free survival of patients in the high diversity group were significantly lower than those in the low diversity group (Figure 3F).
The diversity score calculation method was applied to the validation set (set2), and the analysis showed that the diversity score of some iCCA patients was significantly higher than that of HCC patients (Figure 3H).
04 The expression of VEGF in malignant cells is related to TME recombination
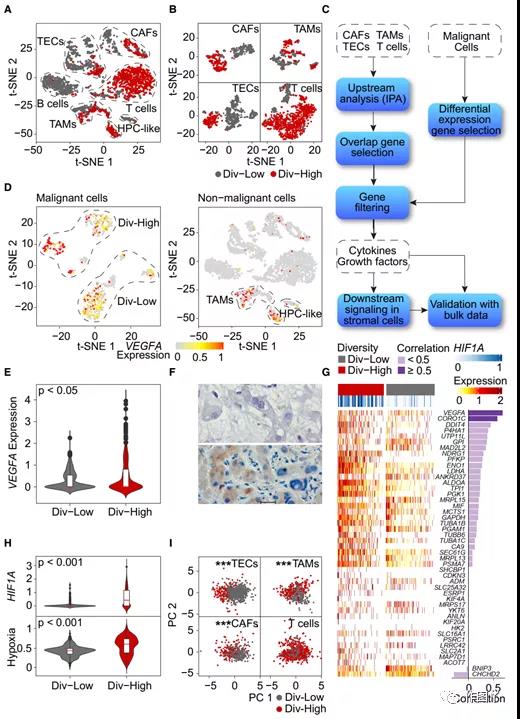
In the validation set 1, the transcriptomes of non-malignant cells from div-high and div-low tumors are different (Figure 4A). When a single cell type was analyzed, the difference was further obvious (Figure 4B), suggesting that TME may be related to tumor cell diversity.
The article found that VEGFA is mainly expressed in malignant cells rather than non-malignant cells (Figure 4D). In addition, the expression level of VEGFA in div-high tumor cells was significantly higher than that in div-low tumor cells (Figure 4E); and immunohistochemical analysis was used to verify the expression pattern of VEGFA in tumor cells (Figure 4F).
Since VEGFA is the direct target of hypoxia-inducible factor 1-α (HIF1A), and HIF1A is a key factor that induces hypoxia, the team determined that in set 1, div-high tumors are more prone to hypoxia than div-low tumors. The team found that the expression levels of hypoxia-related genes in tumor cells in the div-high group were much higher than those in the div-low group (Figure 4G).
The article performs PCA analysis based on the genes of each non-malignant cell type. It was found that the VEGFA functional agent gene can distinguish between div-high and div-low TECs, TAMs and CAFs, but not T cells (Figure 4I).
05 Div-low tumor T cells have cytolytic activity
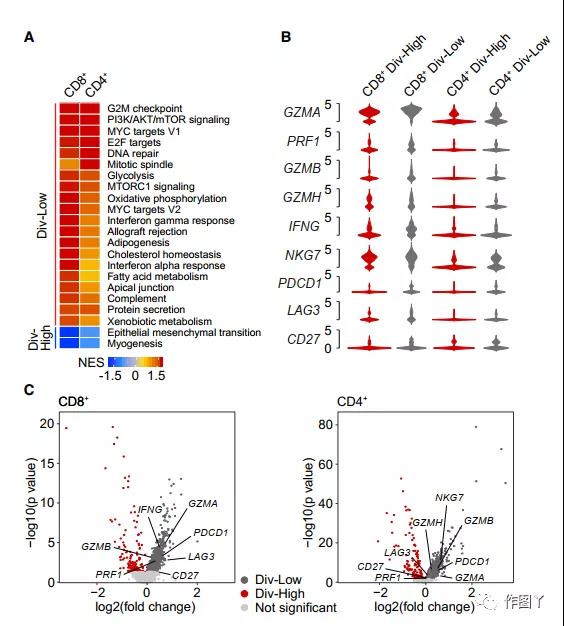
The article uses a clustering algorithm based on Shared Nearest Neighbor (SNN) modular optimization to identify six T cell subtypes (Figure 5A).
The results showed that the branches of CD8+ and CD4+ T cells came from the trajectory tree, where div-high T cells tended to cluster on the trunk, while div-low T cells clustered on the branches (Figure 5B).

This article conducts gene set enrichment analysis based on signaling pathway genes. The enrichment pathways between CD8+ and CD4+ T cells are basically the same (Figure 6A).
In addition, T cells are enriched in the interferon-a (IFN-a)/IFN-g response and MYC activation pathway, as well as the cell proliferation (G2/M checkpoint and E2F target) pathway, suggesting div-low tumor T cells May have cytolytic activity.
Compared with div-high tumors, cytotoxicity-related genes (such as GZMA, GZMB, GZMH and PRF1) from div-low tumor CD8+ and CD4+ T cells were highly expressed (Figure 6B and 6C).
Summary: The diversity of tumor cells drives the reorganization of the liver cancer microenvironment
Liver cancer is the second most deadly malignant tumor in the world, mainly including hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (iCCA).
However, most patients with HCC and iCCA have limited response to molecular targeted therapies.
This article develops a method to measure tumor cell diversity and proves that tumor transcriptome diversity is related to the overall survival of HCC and iCCA patients treated with immune checkpoint inhibitors.
This work also found that TME polarization, including T cell dysfunction, is related to the increased expression of tumor-derived VEGF.
These results indicate the utility of the transcriptome diversity score in predicting the response of immunotherapy, and provide a theoretical basis for the combination of immune checkpoint blockade and antivascular therapy to improve the efficacy.
The diversity of tumor cells drives the reorganization of the liver cancer microenvironment
(source:internet, reference only)
Disclaimer of medicaltrend.org
Important Note: The information provided is for informational purposes only and should not be considered as medical advice.