The relationship between transcription-associated mutations and topoisomerase activity remains unknown
- Normal Liver Cells Found to Promote Cancer Metastasis to the Liver
- Nearly 80% Complete Remission: Breakthrough in ADC Anti-Tumor Treatment
- Vaccination Against Common Diseases May Prevent Dementia!
- New Alzheimer’s Disease (AD) Diagnosis and Staging Criteria
- Breakthrough in Alzheimer’s Disease: New Nasal Spray Halts Cognitive Decline by Targeting Toxic Protein
- Can the Tap Water at the Paris Olympics be Drunk Directly?
The relationship between transcription-associated mutations and topoisomerase activity remains unknown
- Should China be held legally responsible for the US’s $18 trillion COVID losses?
- CT Radiation Exposure Linked to Blood Cancer in Children and Adolescents
- FDA has mandated a top-level black box warning for all marketed CAR-T therapies
- Can people with high blood pressure eat peanuts?
- What is the difference between dopamine and dobutamine?
- How long can the patient live after heart stent surgery?
The relationship between transcription-associated mutations and topoisomerase activity remains unknown
There are diverse mutational processes in cells, and thus cells have multiple strategies to ensure the integrity of the genome, such as high-fidelity DNA replication and DNA repair processes to combat exogenous and endogenous DNA damage [1,2 ] ] .
Regions that target DNA repair during transcription express genes, reducing their mutation rate after DNA damage.
Although this targeted repair mechanism exists, the transcription process itself in microorganisms induces mutations, a phenomenon known as transcription -associated mutagenesis ( TAM ) [3,4] .
In yeast, the topoisomerase Top1 is the major source of TAM and results in a unique 2-5 bp transcription-dependent deletion of tandem repeats.
In general, topoisomerases are considered to protect the stability of the genome by reducing the pressure on the three-dimensional structure of the genome, and are the key to genome maintenance.
Therefore, the relationship between transcription-associated mutations and topoisomerase activity remains unknown.
On February 9, 2022, the research group of Andrew P. Jackson of the University of Edinburgh, the research group of Martin S. Taylor , and Martin AM Reijns (the first author) jointly published the article Signatures of TOP1 transcription-associated mutagenesis in cancer and germline in Nature .
Topoisomerase-mediated transcription-related mutations were found and associated with cancer as well as in germline cells.

According to the classification of cancer somatic mutations, 2-5 bp deletions are called ID4, and usually a single repeat unit is lost in short repeats. In addition to short tandem repeat deletions of less than five repeats, ID4 also includes microhomology deletions, especially single-nucleotide deletions of 2 bp.
However, these deletions are different from the known mutational mechanisms such as replication slippage and non-homologous or microhomologous end joining.
The authors found that these deletions have certain similarities with the transcription-related mutations induced by the topoisomerase Top1 in yeast ( Figure 1) .
Therefore, the authors reanalyzed published genome-wide mutation accumulation experiments in yeast, using an RNase H2/RER-deficient line that is particularly sensitive to Top1-TAM, which accumulates large amounts of ribonucleotides in the genome.
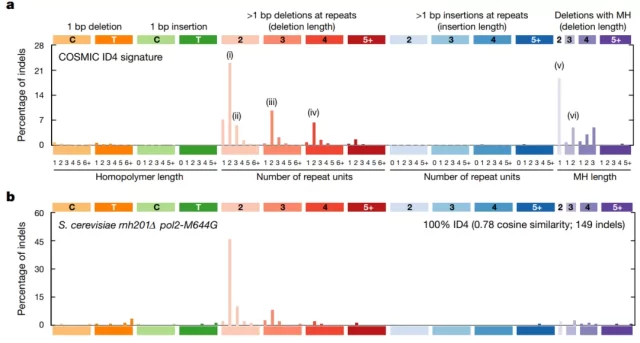 Figure 1 The signature of somatic ID4 mutations in cancer is similar to topoisomerase-induced transcription-related mutations in yeast
Figure 1 The signature of somatic ID4 mutations in cancer is similar to topoisomerase-induced transcription-related mutations in yeast
To assess whether transcription-induced mutations trigger indel formation in human cells, the authors developed a reporter gene that can detect mutational events induced by TOP1 activity in mammals with sufficient sensitivity and specificity.
This reporter gene system is similar to the method inspired by Traffic Light reporter gene analysis [5] , using two selections in a single transcription unit, in the absence of mutations, the hygromycin resistance gene HygroR is expressed; while in the genome there are 2 bp When the hygromycin resistance gene HygroR is not expressed, NeoR is expressed, allowing double selectivity to detect genomic indels (Figure 2) .
The authors inserted this reporter gene into the yeast genome to assess the mutation rate of strains in the absence of RER and/or Top1.
The authors found that the mutation rate was significantly higher in RNase H2/RER-deficient lines compared with wild-type, but the increased mutation frequency was abolished when the Top1 deletion was also introduced.
Therefore, it can be concluded that there are indeed Top1-dependent transcription-related mutations in the yeast genome.
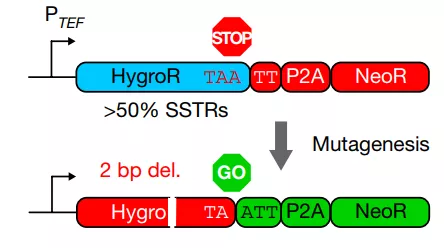 Figure 2 Working model of the “traffic light” reporter gene
Figure 2 Working model of the “traffic light” reporter gene
Are topoisomerase-dependent transcription-related mutations conserved in human cells? To this end, the authors used a similar ‘traffic light’ reporter system in human cells, except that NeoR was replaced by a PuroR gene expressed from the mammalian ubiquitous CAG promoter and antibiotics that allowed rapid selection in mammalian cells.
The authors found that TOP1-induced somatic mutations are conserved across different tissues and cellular contexts, and show that this process can be detected in the context of cancer.
Further, the authors hope to further characterize topoisomerase-induced transcription-related mutations.
The authors focused their attention on 2bp deletions, which occurred more frequently. By analyzing the sequence features surrounding the deletion, the authors found that at least one thymine was deleted in the deletion, because the topoisomerase TOP1 tends to cleave the upstream thymine directly at the phosphodiester bond (Fig. 3) .
Ultimately, the authors determined that the identified sequence was the TNT motif, which is not common across the genome but is present in both short tandem repeat deletions and microhomology deletions.
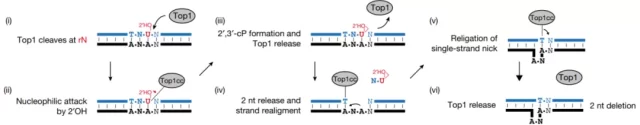 Figure 3 Topoisomerase TOP1 induces transcription-related mutations
Figure 3 Topoisomerase TOP1 induces transcription-related mutations
Because deletions are also present at multiple loci in human cancers, the authors examined the signature of mutations induced by the topoisomerase TOP1 in cancers.
The authors found that in chronic lymphocytic leukemia, topoisomerase TOP1-induced transcription-related mutations are indeed present and enriched in RNase H2-deficient tumors, with a significant frequency of 2-5bp deletions with increased TOP1 activity Increase.
This suggests that TOP1 activity and transcriptional levels also correlate with deletion mutation frequency in cancer cells.
The relevance of TOP1-mediated mutations to other cancers confirms that the ID4-TOP1 mutational signature is transcriptionally relevant and supports the occurrence of transcription-related mutations in humans.
Additionally, TOP1 is ubiquitously expressed in the germline, and the authors reasoned that it may lead to germline and somatic mutations.
Through the analysis of the whole-genome sequencing database of the father-mother-child trio, it was found that most of the gene deletions and deletions were consistent with the ID4-TOP1 signature.
Analysis of 2 bp deletions showed that most deletions occurred in short tandem repeat deletions as well as in regions of microhomology deletions. Thus, the ID4-TOP1 mutational signature is also present in human germ cells, implying that TOP1-induced recombination mutation is an important mutational process in mammalian cells.
Overall, this work found that the topoisomerase TOP1, which has long been considered to maintain the stability of the three-dimensional structure of the genome, induces ID4-TOP1 type deletion mutations in human somatic cells and germ cells, and the process is a transcription-related mutation Process.
The finding could help further research into cancer and the source of the mutation in germ cells.
Reference:
https://doi.org/10.1038/s41586-022-04403-y
The relationship between transcription-associated mutations and topoisomerase activity remains unknown
(source:internet, reference only)
Disclaimer of medicaltrend.org
Important Note: The information provided is for informational purposes only and should not be considered as medical advice.



