Nature Reviews Microbiology | Diversity and evolution of animal virome
- Oregon Reverses Course: From Decriminalization to Recriminalization of Drug Possession
- Why Lecanemab’s Adoption Faces an Uphill Battle in US?
- Yogurt and High LDL Cholesterol: Can You Still Enjoy It?
- WHO Releases Global Influenza Vaccine Market Study in 2024
- HIV Infections Linked to Unlicensed Spa’s Vampire Facial Treatments
- A Single US$2.15-Million Injection to Block 90% of Cancer Cell Formation
Nature Reviews Microbiology | Diversity and evolution of animal virome
- Was COVID virus leaked from the Chinese WIV lab?
- HIV Cure Research: New Study Links Viral DNA Levels to Spontaneous Control
- FDA has mandated a top-level black box warning for all marketed CAR-T therapies
- Can people with high blood pressure eat peanuts?
- What is the difference between dopamine and dobutamine?
- How long can the patient live after heart stent surgery?
Nature Reviews Microbiology | Diversity and evolution of animal virome

The global pandemic of 2019-nCoV makes the significance of studying virus evolution and virus ecology even more important.
In fact, viruses are the most diverse and abundant life forms on earth. They can infect a variety of life forms from lower to higher organisms, regularly spread across species, and may cause serious diseases.
Although the widespread use of next-generation sequencing (mNGS) methods has greatly expanded our knowledge of the diversity of the entire virosphere, our current knowledge is still very limited compared to the viruses that exist throughout the animal kingdom. less part,
Edward Holmes, a well-known virologist and evolutionist from the University of Sydney, Australia, et al. reviewed the main methods currently used for virome research, our understanding and limitations of the structure, diversity and evolution of animal viromes, and how to improve them.
A good understanding of cross-species transmission events, the risk of animal-to-human transmission, and whether evolutionary processes in hosts influence the evolutionary diversity of viruses that infect those hosts.
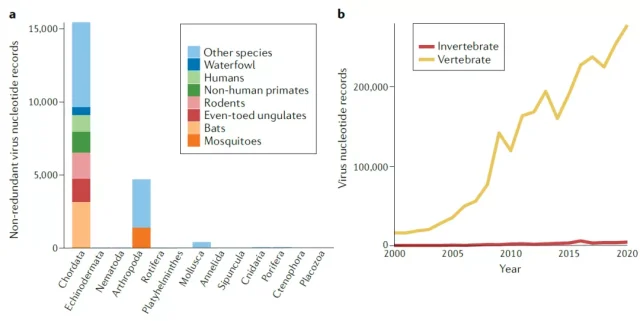 Figure 1. Virus sequence records at different animal taxonomic levels in the NCBI database
Figure 1. Virus sequence records at different animal taxonomic levels in the NCBI database
The biggest problem currently faced in the study of virus diversity is the bias of target virus and sampling.
Due to previous cognition, people generally tend to study known important viruses, especially those that can cause human and important animal-related diseases.
At the same time, host-range studies also tend to focus on some “important” animals, such as chordate bats, rodents, and common invertebrate pathogen carriers-mosquitoes, which makes the diversity of viruses in the database more informative. Focusing on these species makes them “more prominent”.
However, this does not mean that these viruses are lacking in other species.
For example, the authors illustrate that coronaviruses, hepatitis D virus, and influenza viruses—types of viruses often thought to infect humans or common hosts such as mammals and birds—are also found in fish, amphibians, and invertebrates.
This implies that the current research has a large bias and cannot well reflect the diversity and evolutionary relationship of the entire virus in the ecosystem.
And the profound evolutionary history of these hosts also implies that these viruses may also have a long evolutionary history, and their diversity and host range may be far more complex than currently recognized.
Due to the strictly host-parasitic nature of the virus, it is a key question to what extent the evolution, population expansion and extinction of the host, changes in adaptive immunity in history, etc. will affect the diversity and evolutionary relationship of the virus. For example, some viruses occur with the host.
Changes in evolutionary characteristics may lead to the occurrence of diseases.
Although the rate and mechanism of demise and production of viral strains remains unknown, the authors speculate that the reduction in host populations may cause some genetic drift in the virus.
Therefore, studying the taxonomic evolutionary relationship of viruses in different animals on a larger scale and how viruses change with host changes may provide clues for understanding the evolution of virus diversity.
 Figure 2. Virus species and species evolutionary relationships found in different species classifications in the database
Figure 2. Virus species and species evolutionary relationships found in different species classifications in the database
Most human-infected viruses come from animals (zoonoses events), including HIV, SARS-CoV2, etc.
How to analyze the co-evolutionary relationship between a large number of viruses and animals from a more profound historical perspective, including cross-species transmission in evolutionary history Or virus demise events, and figuring out the key factors that influence zoonoses events over a shorter period of time is crucial for us to judge and understand naturally occurring virus cross-species transmission events.
The author also cites a variety of factors in recent times, including animal trafficking, transportation, captivity, climate change, the expansion of human activity areas, and the reduction of forest and animal habitats that have increased human-wildlife contact, while animals Mutual contact between populations themselves due to habitat loss also facilitates gene exchange and the rate of evolution among viruses carried by animals. These factors can alter the diversity of hosts and viruses, and may bring potential risks of cross-species transmission.
The review also suggests that although large-scale mNGS analysis can expand our knowledge of virus diversity in animals and allow for cross-species risk assessment.
However, pure viral genome evolution information cannot accurately determine whether a virus has the risk of cross-species transmission, and a large amount of sufficient experimental evidence is still needed to answer this question.
For example, although bats carry a large number of virus types related to human pathogenic viruses, considering the continuous analysis of viromes in animals, viruses that can be transmitted across species still account for only a very small part.
At the same time, the use of antigen/antibody-based virus screening methods (VirScan) can be used as a supplement to the assessment of viruses with potential transmission risk.
Finally, how to accurately and carefully interpret large-scale virus data is also crucial, such as differences in virus host and geographical distribution caused by animal diets, contamination by multiple laboratory reagents, and the development of new organisms for unknown viruses with farther genetic relationships. Information annotation methods, etc. (such as conservation based on protein structure).
The article suggests that for animal virus library research, the method of meta-transcriptomics can be used, which has a good effect on RNA viruses with the highest diversity.
However, how to efficiently integrate metagenomic and metatranscriptome methods for different scenarios, and how to optimize the removal of host backgrounds and virus-specific enrichment methods also requires more research.

Figure 3. Metagenome sequencing and analysis workflow for virus discovery
Reference:
https://www.nature.com/articles/s41579-021-00665-x
Nature Reviews Microbiology | Diversity and evolution of animal virome
(source:internet, reference only)
Disclaimer of medicaltrend.org
Important Note: The information provided is for informational purposes only and should not be considered as medical advice.



