Knowledge: Learn about various COVID-19 mutants from Alpha to Lambda
- Why Botulinum Toxin Reigns as One of the Deadliest Poisons?
- FDA Approves Pfizer’s One-Time Gene Therapy for Hemophilia B: $3.5 Million per Dose
- Aspirin: Study Finds Greater Benefits for These Colorectal Cancer Patients
- Cancer Can Occur Without Genetic Mutations?
- Statins Lower Blood Lipids: How Long is a Course?
- Warning: Smartwatch Blood Sugar Measurement Deemed Dangerous
Knowledge: Learn about various COVID-19 mutants from Alpha (α)to Lambda (λ)
Knowledge: Learn about various COVID-19 mutants from Alpha to Lambda. Since the outbreak of the SARS-CoV-2 pandemic, many variants have appeared. The most recent outbreak in many countries is the Delta mutant strain.
At the same time, the Lambda mutant strain in South America is spreading rapidly and has spread to more than forty countries around the world. On August 10, according to a report in the “Medical News” magazine, The research team of the New York Health Department in the United States judged that the fatality rate of the new coronavirus variant Iota (B.1.526) found in New York has increased significantly.
What are the similarities and differences between these mutants?
Can the current vaccine still work?
Mutant resistance and infectivity changes
Pseudovirus neutralization experiment to verify the neutralization resistance of the mutant strain to BNT162b2 (Pfizer-BioNTech), mRNA-1273 (moderna), Ad26.COV2.S (Johnson & Johnson) vaccine.
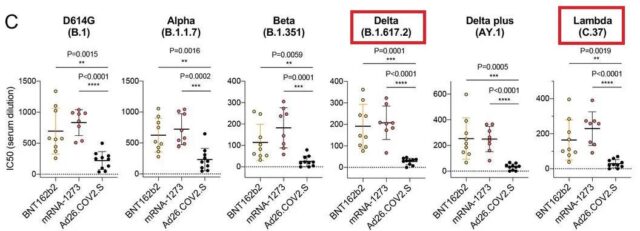
The ordinate is the neutralizing antibody titer (IC50). Each small dot in the figure above represents the result of one neutralization test, and the short horizontal line in the middle of the small dot is the median of several neutralization test results. The higher the position of the dash, the better the neutralizing effect of the neutralizing antibody produced by the vaccine on the strain. You can get a preliminary understanding of the immune escape ability of the corresponding strains by comparing the positions of the dashes of different antibodies.
As shown in the figure, the Lambda strain is more resistant to vaccines than the Delta strain.
Pseudovirus neutralization experiment to verify the neutralization resistance of the mutant strain to Kexing vaccine.
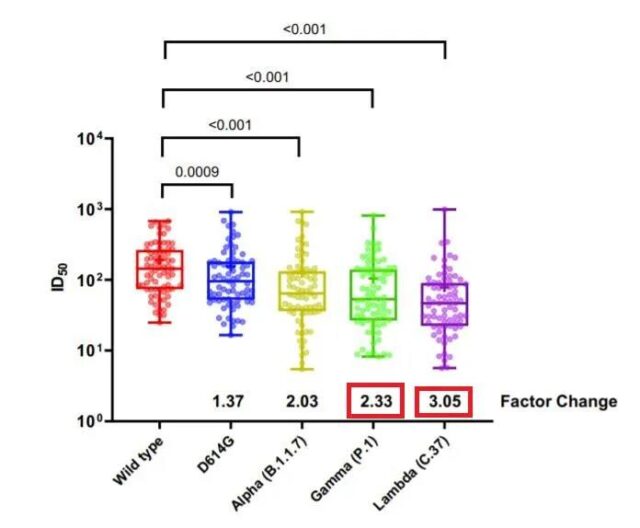
The principle is similar to the above picture, and the height of the dash can be compared. The number below is the reduction factor of antibody neutralization ability. Lambda’s 3.05 means that the neutralizing antibody produced after inoculation with Kexing vaccine has a 3.05 times lower effect on lambda compared to the Wuhan strain.
The reason why Delta can take the world by storm is because the infectivity surpasses Alpha, and the infectivity of Lambda also surpasses alpha.
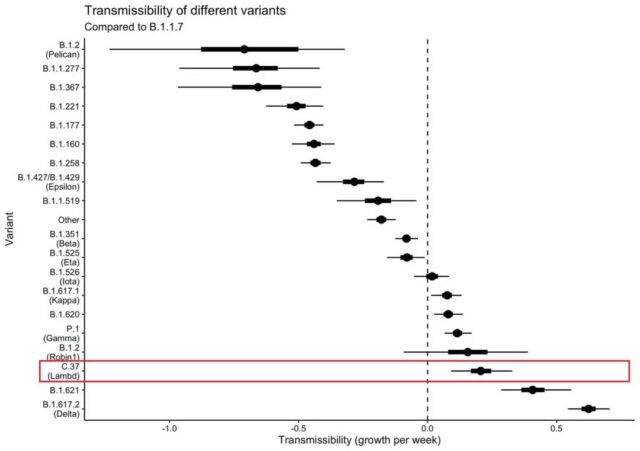
Metadata analysis of the growth rate of the prevalence of various virus strains in 42 countries around the world. The benchmark (black dotted line) in the middle represents Alpha, and the other strains are represented by black dots. The black line above the black dot represents the confidence interval. The closer the black dot is to the right, the more infectious the relative alpha. As shown in the figure, Delta has the first infectivity, B.1.621 has the second infectivity, and Lambda is third.
Introduction to the COVID-19 mutant
The Centers for Disease Control and Prevention and the World Health Organization have established a classification system to distinguish the emerging SARS-CoV-2 variants: Variants of Concern (VOCs) and Variants of interest (VOIs). Such as Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1) and Delta (B.1.617.2), which are all related to transmission and increased virulence and are classified as VOCs; such as Eta (B.1.525), Iota (B.1.526), Kappa (B.1.617.1) and Zeta (P.2) are listed as VOIs.
SARS-CoV-2 Variants of Concern (VOCs)
Alpha (B.1.1.7 lineage)
Alpha (B.1.1.7) mutant strain is a new mutant reported in the UK at the end of December 2020. This strain spread rapidly at the end of 2020. At the beginning of January 2021, it only accounted for 0.2% of the COVID-19 infection cases in the United States in the past two weeks; at the end of February, the proportion rose to 11.4%; by April, the proportion rose to 66%; to It has risen to 72.4% on May 8, becoming the mainstream mutant strain.
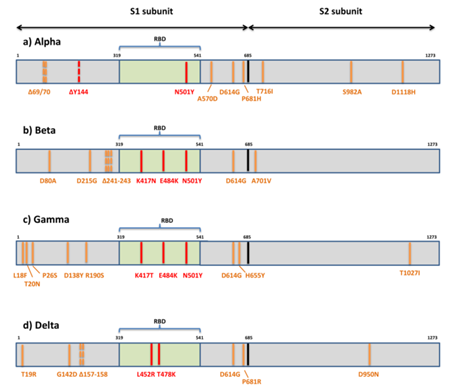
The Alpha strain contains 17 mutations, of which 8 mutations (Δ69-70 deletion, Δ144 deletion, N501Y, A570D, P681H, T716I, S982A, D1118H) are located in Spike protein (S protein, a key protein for viral invasion). The N501Y mutation shows that the Spike protein has an increased affinity for the ACE2 receptor, which enhances the attachment of the virus and subsequent entry into the host cell.
Beta(B.1.351 lineage)
Beta (B.1.351) mutant strain, a new variant reported by Tegally et al. The virus caused a second wave of new coronavirus infections in Nelson Mandela Bay, South Africa in October 2020.
The Spike protein of the Beta strain contains 9 mutations (L18F, D80A, D215G, R246I, K417N, E484K, N501Y, D614G, A701V), of which 3 mutations (K417N, E484K, N501Y) are located in the RBD region (the S protein is affected by ACE2). The body binds to the key area), which improves the binding affinity to the ACE2 receptor. According to reports, this variant has a higher risk of transmission and reduces the neutralizing effect of monoclonal antibody treatment, convalescent serum, and serum after vaccination.
Gamma (P.1 lineage)
The Gamma (P.1) mutant strain was confirmed in Brazil in December 2020 and was first discovered in the United States in January 2021. According to the latest epidemiological information released by WHO on March 30, 2021, the variant has spread to 45 countries.
The Spike protein of the Gamma strain has 10 mutations (L18F, T20N, P26S, D138Y, R190S, H655Y, T1027IV1176, K417T, E484K and N501Y), and three mutations (L18F, K417T, E484K) are located in the RBD region, similar to the Beta strain. Importantly, this variant may reduce the neutralizing effect of monoclonal antibody therapy, convalescent serum, and post-vaccination serum.
Delta(B.1.617.2 lineage)
The Delta (B.1.617.2) mutant strain, originally discovered in India in December 2020, was responsible for the second wave of fatal COVID-19 infections in India in April 2021. In the United States, this variant was first discovered in March 2021 and will become the most important SARS-CoV-2 strain in the United States in the coming weeks. In July 2021, the COVID-19 epidemic broke out in Nanjing. The genome sequencing results of the cases in the epidemic showed that the strain was a delta mutant.
The Spike protein of the Delta strain contains 10 mutations (T19R, (G142D*), 156del, 157del, R158G, L452R, T478K, D614G, P681R, D950N), which has stronger adaptability to the human body, faster transmission speed, and viral load Higher, longer treatment time, and more likely to develop into severe illness. In addition, the Delta mutant strain can evade the neutralizing ability of the neutralizing antibody produced by the immune system. The study released by Moderna shows that compared with the wild-type new coronavirus strain containing the D614G mutation, the serum of mRNA-1273 COVID-19 vaccine inoculators has the Delta mutation The neutralizing ability of the strain was reduced by 2.1 times.
SARS-CoV-2 Variants of interest (VOIs)
Epsilon (B.1.427 and B.1.429)
Epsilon (B.1.427 and B.1.429) mutant strains appeared in the United States around June 2020. From September 1, 2020 to January 29, 2021, the number of sequenced cases increased from 0% to 50%. Compared with wild-type epidemic strains, the dissemination has increased by 18.6-24%.
The Epsilon strain has the following mutations (B.1.427: L452R, D614G; B.1.429: S13I, W152C, L452R, D614G). Due to its increased transmission capacity, the US Centers for Disease Control and Prevention listed this strain as VOI (Variants of Interest).
Zeta (P.2)
Zeta (P.2) mutant strain was first discovered in Brazil in April 2020. Its Spike protein has key mutations (L18F; T20N; P26S; F157L; E484K; D614G; S929I; V1176F). Because this variant may reduce the neutralizing effect of antibody therapy and vaccine serum, it has been approved by the World Health Organization and the United States for Disease Control. The Prevention Center is listed as VOI.
Eta (B.1.525) and Lota (B.1.526)
Eta (B.1.525) and Lota (B.1.526) mutant strains were first discovered in New York in November 2020. Since this mutant strain may reduce the neutralization effect of antibody therapy and vaccine serum, CDC and WHO list them as VOI. On August 10, according to a report in the “Medical News” magazine, the research team of the New York Health Department judged that the death rate of the new coronavirus variant Iota (B.1.526) found in New York has increased significantly. During the study period, compared with other COVID-19 variant strains, the mortality of infected people aged 45-65, 65-74, and 75 years old increased by 46%, 82%, and 62%, respectively.
Its Spike protein has key mutations (B.1.525: A67V, Δ69/70, Δ144, E484K, D614G, Q677H, F888L; B.1.526: (L5F*), T95I, D253G, (S477N*), (E484K*), D614G, (A701V*)).
Theta (P.3)
Theta (P.3) mutant strain, whose Spike protein carries key mutations (141-143 deletion E484K; N501Y; P681H), and was first discovered in the Philippines and Japan in February 2021. It used to be VOI. Later, due to the decreasing number of cases of these mutant strains, the impact on the epidemic became lower and lower. Now it has been downgraded to “mutant strains requiring further monitoring” (alerts for further monitoring)
Kappa (B.1.617.1)
Kappa (B.1.617.1) mutant strains carrying key mutations ((T95I), G142D, E154K, L452R, E484Q, D614G, P681R and Q1071H) were first discovered in India in December 2020. It belongs to one of the subspecies of India mutant virus along with the Delta mutant. The significant mutation of Spike protein makes Kappa virus, like Delta, more likely to infect cells and avoid the immune system’s antibody response.
Lambda(C.37)
Lambda (C.37) mutant strain was first discovered in Peru. When the medical community first noticed Lambda, only 1 case could be detected from 200 samples. By March 2021, about 50% of the COVID-19 cases in Lima, the capital of Peru, originated from this variant. WHO data shows that between May and June this year, 82% of new cases in Peru were caused by the Lambda variant, and the per capita death rate of COVID-19 soared to the highest in the world. At present, the mutant strain has spread in dozens of South American countries including Peru, Chile, Argentina, and Ecuador. In the United Kingdom and other countries, infections with the Lambda mutant strain have also been found.
Lambda strain has many mutations in Spike protein, mainly including G75V, T76I, del247/253, L452Q, F490S, D614G and T859N. Among them, RSYLTPGD246-253N, L452Q and F490S mutations lead to increased immune resistance; and T76I and L452Q mutations lead to increased infectivity. This allows the Lambda strain to have the ability to increase infectivity and immune escape at the same time, thus gaining the potential for large-scale spread.
From the pseudovirus neutralization experiment, it can be seen that not all vaccines have a good protective effect on the mutant strains, and in principle, the vaccine will not achieve 100% protection, and individual immune responses must be different. Therefore, it is necessary to evaluate the valence of the protection rate of the mutant strain of the vaccine to guide vaccination, epidemic prevention and control, and disease treatment.
Pseudovirus system and immune blocking methods are commonly used to evaluate vaccine-induced neutralizing antibodies.
At the end of the article, you can see the list of pseudoviruses, Spike mutant proteins, RBD mutant proteins and other products provided by Nearshore Protein.
COVID-19 vaccine evaluation method
False virus infection verification
Neutralizing antibodies can prevent viruses from infecting cells. Add neutralizing antibodies (serum/neutralizing antibody positive control after vaccine injection) to the virus-infected cell system to detect the reduction of infected cells and calculate the blocking effect.
Due to the high infectivity and pathogenicity of the new coronavirus, the use of live new coronavirus for infection verification must be carried out in a biosafety level 3 or above laboratory, which is limited by laboratory conditions and the source of the virus. In addition, due to the different strains, culture conditions, and evaluation criteria of the results, there are often some differences in the results of live virus detection in different laboratories. In terms of safety and maneuverability, it is more convenient to use pseudoviruses, and pseudoviruses are easy to achieve standardization, which is conducive to evaluating the antiviral effects of vaccines in preclinical and clinical stages. By comparing different mutant pseudovirus strains to the immunized serum samples for neutralizing antibody detection, the neutralizing effect of the vaccine on the mutant strain can be effectively evaluated.
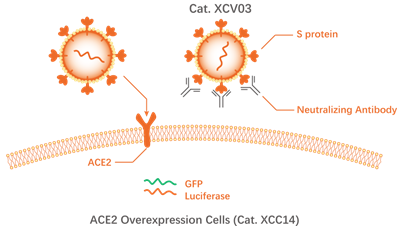 Neutralizing antibodies inhibit the infection of cells by the new coronavirus pseudovirus (source: novoprotein)
Neutralizing antibodies inhibit the infection of cells by the new coronavirus pseudovirus (source: novoprotein)
Pseudovirus detection system: Novoprotein Cat.XCV03 has no self-replication ability. It expresses full-length S protein on its surface and contains green fluorescent protein (GFP) and luciferase (Luciferase) reporter genes, which can specifically infect Cells expressing ACE-2 (Novoprotein Cat. XCC14). The cells infected by the virus express GFP and Luciferase and emit fluorescence, so that the results can be easily observed and detected by a chemiluminescence microplate reader to realize quantitative analysis. Pseudovirus assays can be further optimized to achieve high-throughput detection and standardized detection. By replacing the S protein expression plasmid (protein coding gene), the cross-neutralization effect of anti-S protein antibodies against different mutant strains can be studied.
Sum up:
1. The false virus detection system has high virus titer, good detection accuracy and repeatability;
2. The pseudovirus contains a fluorescent reporter gene, which is conducive to quantification and standardization;
3. Pseudovirus has no self-replication ability and only infects cells expressing ACE-2, which is safer.
Immune blocking verification
The virus infection verification method is to verify the effectiveness of neutralizing antibodies at the cellular level, and is suitable for the vaccine development stage. Using the protein of the mutant strain, for the population after mass vaccination, the protective effect of each person’s neutralizing antibody on the mutant strain can be quickly evaluated, and a simpler and faster immune blocking verification method can be used.
Immune blocking verification uses the principle that neutralizing antibodies block the binding of S-RBD protein to ACE-2. After adding neutralizing antibodies, detecting the reduced amount of S-RBD protein bound to ACE-2 is used to calculate the drop of neutralizing antibodies. Spend.
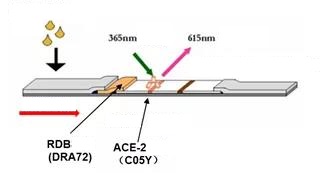
Time-resolved fluorescence immunochromatography detection neutralizing antibody competitively inhibits S-RBD binding to ACE-2
Using the same immune blocking principle, ELISA or lateral chromatography colloidal gold method can also be used to detect neutralizing antibodies.
ELISA, neutralizing antibody blocks the binding of S protein to ACE-2, IC50 is 25ng/ml (Source: www.novoprotein.com.cn)
Sum up:
1. The operation is simple, the detection time is short (<30min), and it can be used for on-site screening and large-scale detection;
2. Low cost and high cost performance, which is conducive to large-scale testing;
3. The results are intuitive and can be applied to rapid detection after vaccination.
T cell immune response assessment
Some current studies have shown that in the fight against COVID-19, the induction of neutralizing antibody responses alone cannot fully protect the body, and the immune response of T cells also plays a very important role. Studies have shown that neutralizing antibodies, helper T cells, and killer T cells can be detected in all fully recovered individuals.
Generally speaking, people with extensive and well-coordinated immune responses have milder, special, and intense illnesses. The new coronavirus-specific T cell response is a precursor to milder symptoms. Some mild patients will trigger a strong memory T cell response even if the virus-specific antibody response is not detected. People over 65 years of age are more likely to have poor T cell responses and uncoordinated immune responses, and therefore a more serious or fatal COVID-19.
The large part of the susceptibility of the elderly to COVID-19 seems to be a weaker adaptive immune response, which may be due to the fact that the elderly have fewer initial T cells. In addition, memory T cells after vaccination will continue to protect the body, so it is very important to evaluate the T cell immune response induced by the vaccine.
Studies have found that new coronavirus N protein-specific T cells have also been detected in people who have not been infected with COVID-19, and that the specific T cells in SARS recovered patients have strong cross-reactivity with SARS-CoV-2. This indicates that the common β-coronavirus infection can induce the human body to produce multi-specific and durable T cell immunity to the structural protein NP of the coronavirus. Therefore, to verify the specific T cells induced by the vaccine, the COVID-19-specific S-trimer Protein (Novoprotein Cat. DRA49) should be used as the main.
To evaluate the T cell immune response triggered by the vaccine, the gold standard is IFN-γ ELISpot to detect the immune response of PBMC (peripheral blood mononuclear cells), that is, the S protein is used to stimulate the PBMC in the same volunteer before and after vaccination, and T cell secretion after vaccination The degree of IFN-γ should be several times before vaccination.
Enzyme-Linked Immunospot Assay (ELISpot) can detect antibody-secreting cells or cytokine-secreting cells at the single-cell level. As a cytokine secreted by immunocompetent cells, IFN-γ plays an important role in inducing antiviral immunity, including activation of cytotoxic lymphocytes (CTL) and natural killer cells (NK) cells And phagocytes, etc., and the level of IFN-γ produced by the body after vaccine immunization actually reflects the activity of helper T cells. Therefore, the use of ELISpot to detect the level of IFN-γ is an indirect detection of helper T cell activity.
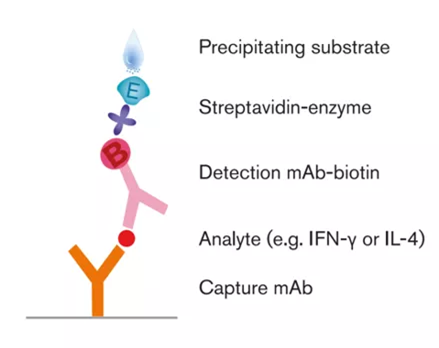 ELISpot detects PBMC secretion of IFN-γ
ELISpot detects PBMC secretion of IFN-γ
IFN-γ ELISpot operation process
1. Coat the bottom of the detection hole with anti-IFN-γ monoclonal antibody;
2. Separate the PBMC of the sample to be tested;
3. Put the PBMC to be tested into the detection hole and add the stimulant S-trimer Protein (Novoprotein Cat.DRA49) at the same time. During the culture period, T lymphocytes that respond to S protein will be activated and begin to secrete specific cytokines IFN-γ, which are simultaneously captured by the monoclonal antibody at the bottom of the plate; cells that do not respond to S protein are not. It is stimulated and does not secrete specific cytokine IFN-γ;
4. Remove the cells, leaving potential “images” of cytokines at the bottom of the plate;
5. Add a biotin-labeled detection antibody, and the detection antibody binds to the cytokine on the “image” to form a sandwich structure of “antibody-antigen-antibody”;
6. Add streptavidin-labeled enzyme solution and combine with the labeled biotin on the detection antibody through streptavidin to form a complex;
7. Adding the chromogenic substrate, under the catalytic decomposition of the enzyme, insoluble pigments are generated, which are deposited on the local membrane to form spots and counted.
Each spot represents a specific T lymphocyte that responds to a specific antigen. By using different mutant strain proteins, the immune response of T cells to the mutant strain can be evaluated, and thus the protective effect of the vaccine on the mutant strain can be evaluated. The number of spots reflects the recognition status of the sample’s cellular immunity: more spots indicate a good immune recognition status, and fewer spots indicate a poor immune recognition status or immune tolerance.
Sum up:
1. ELISpot has high sensitivity. Only one positive cell secreting cytokine can be detected in one million negative cells.
2. ELISpot detects the secretion of a single cell, not the average secretion of a cell population.
3. The cytokines detected by ELISpot do not exist in the normal state of cells, but are secreted by living cells after being stimulated by stimuli during the detection process. The strength and number of spots directly reflect the ability of cells to respond to stimuli and to secrete cytokines.
4. The result is intuitive, one positive cell corresponds to one spot.
(source:internet, reference only)
Disclaimer of medicaltrend.org
Important Note: The information provided is for informational purposes only and should not be considered as medical advice.