The most comprehensive single-cell spatial map of breast cancer so far
- Normal Liver Cells Found to Promote Cancer Metastasis to the Liver
- Nearly 80% Complete Remission: Breakthrough in ADC Anti-Tumor Treatment
- Vaccination Against Common Diseases May Prevent Dementia!
- New Alzheimer’s Disease (AD) Diagnosis and Staging Criteria
- Breakthrough in Alzheimer’s Disease: New Nasal Spray Halts Cognitive Decline by Targeting Toxic Protein
- Can the Tap Water at the Paris Olympics be Drunk Directly?
The most comprehensive single-cell spatial map of breast cancer so far
- Should China be held legally responsible for the US’s $18 trillion COVID losses?
- CT Radiation Exposure Linked to Blood Cancer in Children and Adolescents
- FDA has mandated a top-level black box warning for all marketed CAR-T therapies
- Can people with high blood pressure eat peanuts?
- What is the difference between dopamine and dobutamine?
- How long can the patient live after heart stent surgery?
Nature Sub-Journal: The most comprehensive single-cell spatial map of breast cancer so far
The tumor heterogeneity of breast cancer is extremely large, and these cells with different functions are very important in determining the cause and treatment plan.
Studies have found that tumor-infiltrating lymphocytes (TIL) are biomarkers that respond well to neoadjuvant chemotherapy, while tumor-associated macrophages (TAM) are associated with poor prognosis and are new targets for cancer immunotherapy.
In addition, mesenchymal cells have also become important regulators of drug resistance, driving metastasis and anti-tumor immunity. However, the clear subclass of these cells is currently unclear.
Single-cell RNA sequencing (scRNA-seq) comprehensively and systematically map the cell map of tumors, revealing cell biology, disease etiology, and drug response.
Studies have applied scRNA-seq to human breast cancer to reveal the continuous differentiation status in TILs, the role of tissue-resident lymphocytes in triple-negative breast cancer (TNBC), and the chemotherapy resistance of tumor cells in TNBC.
There is an urgent need for a more detailed high-resolution breast cancer cell atlas to further determine the classification of breast cancer, identify cell heterogeneity and their interactions, and determine cell differentiation events.
On September 6, 2021, the research team of the Garvan Institute of Medical Research in Australia published a research paper entitled: A single-cell and spatially resolved atlas of human breast cancers in the sub-Journal Nature Genetics.
The research developed a single-cell method based on inherent subtype classification (SCSubtype) to reveal the heterogeneity of recurrent tumor cells, providing the most comprehensive picture of breast cancer cells to date.
The study found a new population of PD-L1/PD-L2+ macrophages related to clinical outcomes. Mesenchymal cells are divided into three distinct subgroups.
The spatial transcriptome reveals the interaction between stromal cells and immune cells, and provides unique insights into anti-tumor immune regulation.
The deconvolution of a large breast cancer cohort using single-cell characteristics divides the stages into nine “ecotypes”, which have unique cell composition and clinical results.

First, in order to clarify the cell structure of breast cancer, the research team analyzed three common clinical subtypes (estrogen receptor positive, ER+; human epidermal growth factor receptor 2 positive, HER2+; triple negative breast cancer, using scRNA-seq). TNBC) 26 cases of primary tumors, and identified a type of malignant epithelial cells.
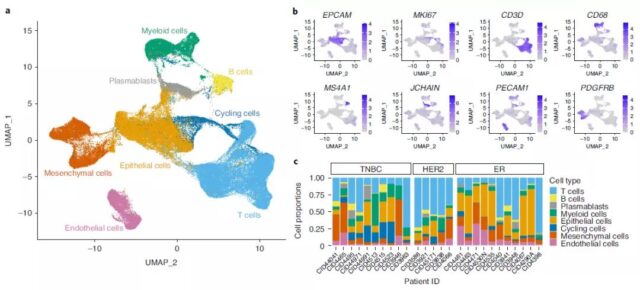
Based on extensive transcriptome analysis using PAM50 gene signatures, breast cancer is divided into five intrinsic molecular subtypes.
However, the function of breast cancer molecular subtypes at the individual cell level is still unclear.
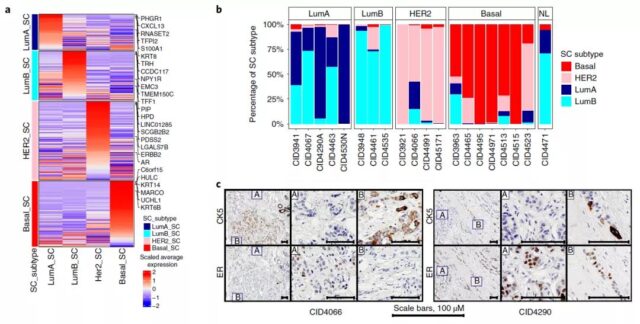
So the research team developed a method compatible with scRNA-seq for intrinsic molecular subtype typing—SCSubtype.
The results show that SCSubtype can reveal cell heterogeneity, correctly identify malignant cells, and has higher specificity than batch and scRNA-seq.
Next, the research team studied the biological pathways that drive intratumoral transcriptional heterogeneity (ITTH) and generated 574 ITTH gene signatures.
These gene characteristics identified seven gene modules (GMs) with different functional characteristics. GMs also revealed the heterogeneity of tumor cells.
In general, the analysis of SCSubtype and GMs provides a complementary new method for ITTH classification and further proves the great heterogeneity of tumor cells.
In order to examine the microenvironment of breast tumors with high resolution, the research team reclassified immune cells and stromal cells, and identified 18 T cells, congenital lymphocyte clusters and 13 myeloid cell clusters in patients, and 5 Three kinds of cancer fibroblast cell clusters, three kinds of endothelial cell clusters and three kinds of parietal cell clusters.
Immune cell clusters are qualitatively different between different clinical subtypes, indicating that the most suitable targeting strategy needs to be formulated according to different clinical subtypes.
They also found similar stromal cell clusters in different breast cancer subtypes and normal breast tissue samples, indicating that stromal cell clusters may be resident cell types undergoing remodeling in the tumor environment.
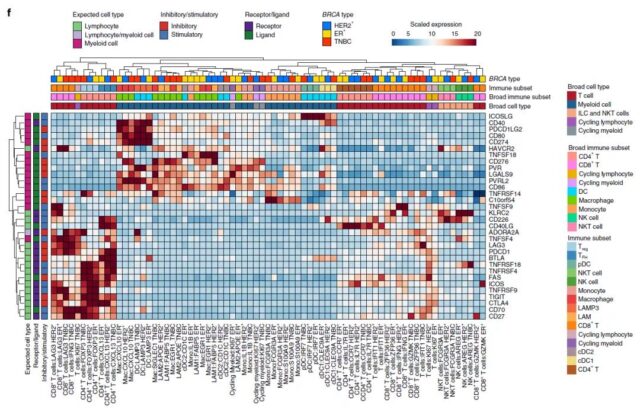
Spatial transcriptome analysis shows that related cell clusters of different clinical subtypes are mutually exclusive in spatial arrangement.
Finally, based on the single-cell results and the large breast cancer cohort, the research team performed a cluster analysis.
The results showed that the nine tumor clusters have similar cell composition, which is called “ecotype”.
These ecotypes are shown to be related to tumor subtypes, SCSubtype cell distribution and the diversity of main cell types, and there are obvious differences in prognosis between ecotypes.
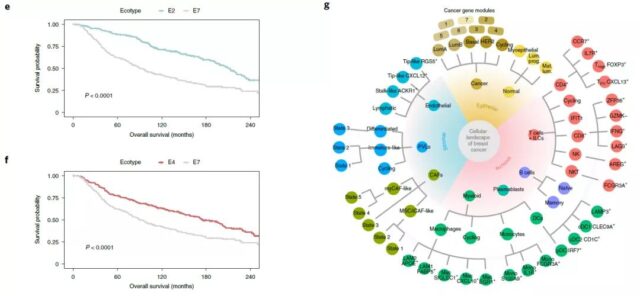
In general, this work defines the cellular atlas of breast cancer at three levels—single-cell RNA sequencing, spatial transcriptome and immunophenotype, revealing a new immunophenotype of breast cancer, and a comprehensive classification of breast cancer. Cell models provide an important basis.
Original link:
https://www.nature.com/articles/s41588-021-00911-1
The most comprehensive single-cell spatial map of breast cancer so far
(source:internet, reference only)
Disclaimer of medicaltrend.org
Important Note: The information provided is for informational purposes only and should not be considered as medical advice.



