Nature Biotechnology: RNA editing has reached a new level!
- Why Botulinum Toxin Reigns as One of the Deadliest Poisons?
- FDA Approves Pfizer’s One-Time Gene Therapy for Hemophilia B: $3.5 Million per Dose
- Aspirin: Study Finds Greater Benefits for These Colorectal Cancer Patients
- Cancer Can Occur Without Genetic Mutations?
- Statins Lower Blood Lipids: How Long is a Course?
- Warning: Smartwatch Blood Sugar Measurement Deemed Dangerous
NatureNature Biotechnology: RNA editing has reached a new level! Accurate and efficient RNA editing without exogenous protein
- Red Yeast Rice Scare Grips Japan: Over 114 Hospitalized and 5 Deaths
- Long COVID Brain Fog: Blood-Brain Barrier Damage and Persistent Inflammation
- FDA has mandated a top-level black box warning for all marketed CAR-T therapies
- Can people with high blood pressure eat peanuts?
- What is the difference between dopamine and dobutamine?
- How long can the patient live after heart stent surgery?
Nature Biotechnology: RNA editing has reached a new level! Accurate and efficient RNA editing without exogenous protein.
The treatment of genetic diseases has always been a difficult problem to be solved.
Fortunately, with the development of human genomics, more and more human genetic disease genes have been discovered one after another.
By changing the genetic sequence of patients with genetic diseases to fundamentally cure human genetic diseases, it has become a promising treatment. method.
Recently, the development of various programmable, site-specific tools for manipulating genetic information has created opportunities to correct disease-causing genetic mutations, such as DNA base editors developed by David Liu et al .
However, it is worth noting that DNA editing may induce heritable and permanent off-target mutations, which greatly limits the application of DNA base editors in humans. In contrast, RNA editing only changes mRNA transiently, avoiding safety and ethical concerns.
Recently, the team of Professor Thorsten Stafforst of the University of Tübingen in Germany published a research paper entitled: CLUSTER guide RNAs enable precise and efficient RNA editing with endogenous ADAR enzymes in vivo in the journal Nature Biotechnology .
This study developed a cluster of guide RNAs , CLUSTER gRNAs , that bind target mRNAs in a multivalent manner without the need for exogenous proteins , utilize endogenous ADAR enzymes to achieve high-precision and high-efficiency editing, and enable targeted editing of previous Sequences inaccessible to gRNA design .
In cell experiments, CLUSTER gRNAs achieved an editing efficiency of up to 45%, and in mouse experiments, the editing efficiency of CLUSTER gRNAs was also as high as 10%.

RNA base editing is a promising option for genome editing. Currently, several RNA editing tools have been designed to target adenosine-inosine (A-to-I) and cytidine-uridine (C-to-U) transitions.
These RNA editing tools require the ectopic expression of an engineered editing enzyme, potentially resulting in substantial off-target editing.
Indeed, ectopic expression of the highly active A-to-I editing enzyme led to lethal consequences in a fragile mouse model of the disease.
Based on this, more and more studies have used endogenous RNA-specific adenosine deaminase (ADAR) to solve the problem of ectopic expression of engineered editing enzymes, but there are still problems such as sequence limitations, low editing efficiency and off-target editing.
As early as October 2016, the team of Prof. Thorsten Stafforst published the related research on RNA editing using endogenous ADAR in Nucleic Acids Research .
The article is titled: Harnessing human ADAR2 for RNA repair – Recoding a PINK1 mutation rescues mitophagy .
The research team designed a special gRNA that contains complements that bind to the target sequence, as well as RNA structural motifs that recruit ADARs.
The design was particularly successful when these gRNAs were used in the form of chemically modified oligonucleotides, yielding between 20-30% editing yields for related transcripts such as STAT1.

Notably, the researchers found that the same oligonucleotide sequences were much less effective when expressed in plasmids.
One possible explanation is that chemically modified oligonucleotides have stronger binding affinity.
Thus, plasmid-borne gRNAs successfully edited endogenous targets via endogenous ADARs when the specificity domain was extended beyond 100 nt.
Thorsten Stafforst referred to these gRNAs as LEAPER gRNAs, however, the LEAPER approach showed substantial off-target editing due to longer gRNAs binding off-target mRNAs more readily.
Therefore, the research team improved it and described the design principles of new CLUSTER gRNAs – including a specificity domain that binds to the target mRNA and a set of 7-20nt Recruitment sequence (RS) , and recruitment sequence (RS) RNA structural motifs of ADAR.
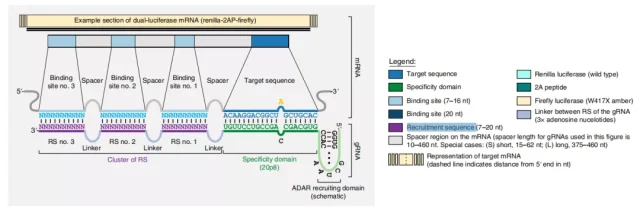 Design principles for CLUSTER gRNAs
Design principles for CLUSTER gRNAs
This CLUSTER design enables genetically encoded gRNAs with a high degree of sequence flexibility, which bind to their target mRNAs in a multivalent manner, enabling high-precision and high-efficiency editing, and the ability to target sequences inaccessible using previous gRNA designs.
 CLUSTER gRNAs enable RNA editing with higher precision and efficiency
CLUSTER gRNAs enable RNA editing with higher precision and efficiency
CLUSTER gRNAs can be genetically encoded and delivered via viral vectors, enabling high-efficiency editing in a variety of cell lines.
In cell culture, CLUSTER gRNAs enable targeted editing of endogenous transcripts with efficiencies as high as 45%.
In vivo, delivery of CLUSTER gRNAs into mouse liver via hydrodynamic tail vein injection (HTVi) resulted in up to 10% of edited reporter gene constructs.
 CLUSTER gRNAs show high RNA editing efficiency in both in vitro and in vivo experiments
CLUSTER gRNAs show high RNA editing efficiency in both in vitro and in vivo experiments
In conclusion, this study developed a clustered guide RNA, CLUSTER gRNAs, that recruited endogenous ADAR enzymes for RNA editing and exhibited higher editing efficiency and fewer off-target effects.
The CLUSTER method opens up new avenues for drug development in the field of RNA base editing!
Reference:
https://www.nature.com/articles/s41587-021-01105-0
Nature Biotechnology: RNA editing has reached a new level!
(source:internet, reference only)
Disclaimer of medicaltrend.org
Important Note: The information provided is for informational purposes only and should not be considered as medical advice.