Breast cancer patients may have chromosomal mutations as early as 6 years old
- Normal Liver Cells Found to Promote Cancer Metastasis to the Liver
- Nearly 80% Complete Remission: Breakthrough in ADC Anti-Tumor Treatment
- Vaccination Against Common Diseases May Prevent Dementia!
- New Alzheimer’s Disease (AD) Diagnosis and Staging Criteria
- Breakthrough in Alzheimer’s Disease: New Nasal Spray Halts Cognitive Decline by Targeting Toxic Protein
- Can the Tap Water at the Paris Olympics be Drunk Directly?
Breast cancer patients may have chromosomal mutations as early as 6 years old
- Should China be held legally responsible for the US’s $18 trillion COVID losses?
- CT Radiation Exposure Linked to Blood Cancer in Children and Adolescents
- FDA has mandated a top-level black box warning for all marketed CAR-T therapies
- Can people with high blood pressure eat peanuts?
- What is the difference between dopamine and dobutamine?
- How long can the patient live after heart stent surgery?
Breast cancer patients may have chromosomal mutations as early as 6 years old.
“Nature”: Scientists have discovered for the first time that breast cancer patients may have common chromosomal rearrangement driver mutations as early as 6 years old.
The research results published by Seiji Ogawa’s team at Kyoto University in “Nature” on July 27, 2023.
They systematically studied the evolutionary history of breast cancer based on human samples and found that the chromosomal rearrangement driver mutation der(1;16) present in 20% of breast cancer patients appeared when the patients were 5.8-16.9 years old; etc. When the patient grew up to 18.1-34.4 years old, driven by der(1;16), the most recent common ancestors (MRCAs) of breast cancer cells appeared [1].
Remarkably, they observed a similar pattern of evolution in breast cancer cases driven by other mutations. Professor Ogawa believes that their discovery will help people understand the occurrence process of breast cancer, and provide new ideas for the prediction, early diagnosis and even prevention of breast cancer .

Screenshot of paper home page
In recent years, a large number of studies have found that those seemingly normal human tissue cells actually carry a large number of cancer-driving mutations [2,3], and the existence of these mutations paved the way for the occurrence of cancer.
However, for a specific cancer type, scientists still know little about when the initial driver mutations that lead to cancer occurred and how they evolved step by step, eventually leading to cancer .
In Professor Ogawa’s opinion, to understand the evolutionary history of cancer, it is necessary to study normal tissues and cancer tissues together.
They set the research object as breast cancer, because breast cancer has become the largest cancer in the world, and the evolution of breast cancer is not enough.
To understand the evolutionary history of breast cancer, an important first task is to estimate the rate of mutation accumulation in normal breast epithelial cells with age.
To this end, they recruited 6 premenopausal breast cancer patients, 9 postmenopausal breast cancer patients and 6 healthy women, collected breast epithelial cells from their normal breast tissue or milk, and then carried out whole genome sequencing.
From the sequencing results, they identified a total of 58,385 single nucleotide variations (SNVs) and 3,955 small insertion/deletion (indels) mutations .
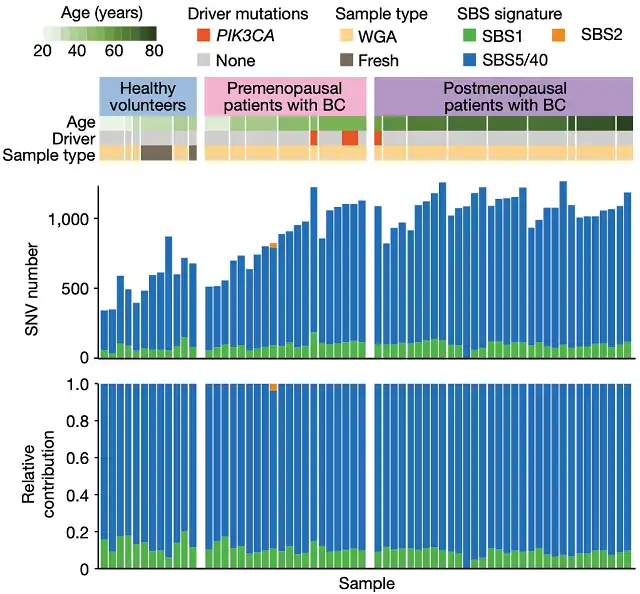
Mutation signatures for all participants
After calculation, Professor Ogawa’s team found that before menopause, the accumulation of SNVs was 19.5 mutations per genome per year, while after menopause it dropped to 8.1 mutations per genome per year ;
the cumulative rate of insertion-deletion mutations also decreased by 45% , from 1.3 per genome per year in premenopause to 0.72 per genome per year in postmenopause .
With this basic data, they can carry out follow-up research.
In order to accurately study the evolutionary history of breast cancer, they have very strict requirements on the research samples.
The pathological samples of breast cancer are required to include not only cancer tissue, but also three or more non-cancerous hyperplasia lesions with a diameter of 3 mm or more.
From 156 pathological samples of breast cancer, they only confirmed that the samples of 5 patients met the requirements (numbers: KU539, KU582, KU779, KU873 and KU957), all of them were in their 40s and not menopausal.
Subsequently, they used laser capture microdissection (LCM) technology to cut 69 research samples with an average diameter of 2.9 mm (0.5-10) from the lesion tissue, and carried out whole genome sequencing.
Among them, 6 were histologically normal lobules, 1 was non-proliferative lesion, 33 were proliferative lesions, 1 was typical lobular carcinoma in situ (LCIS, precancerous lesion), 20 were ductal carcinoma in situ (DCIS), invasive 8 copies of ductal carcinoma (IDC).
Seen from the phylogenetic tree, the five patients only included one or two major clades .
In these lineages, multiple descendants from a common ancestor often acquire their own unique driver gene alterations that evolve cancer or benign breast lesions (BBLs) .
These progeny amplified over the years, and by the time the cancer was diagnosed, they had taken over a large area of the breast.
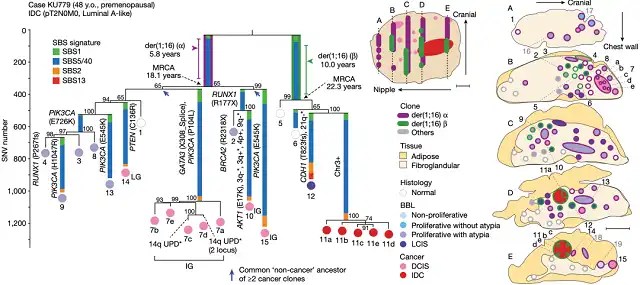
48-year-old KU779 research data
After comparing the research data, a chromosomal rearrangement driver mutation called der(1;16) attracted the attention of Prof. Ogawa’s team, because in 4 out of 5 patients, the most recent common ancestor (MRCA) of each lineage carried There is der(1;16).
der(1;16) is an abnormal chromosome produced by the unbalanced translocation between the q arm of chromosome 1 and the p arm of chromosome 16 near the center point , and it is also a characteristic chromosome of Luminal A breast cancer.
The test results showed that der(1;16)-positive clones were widely distributed in the breast, spanning an average area of 62 mm, and showed different histological features, including noncancerous hyperplastic lesions, LCIS, DCIS, and IDC . It is suggested that these good and evil organizations have a common ancestor.
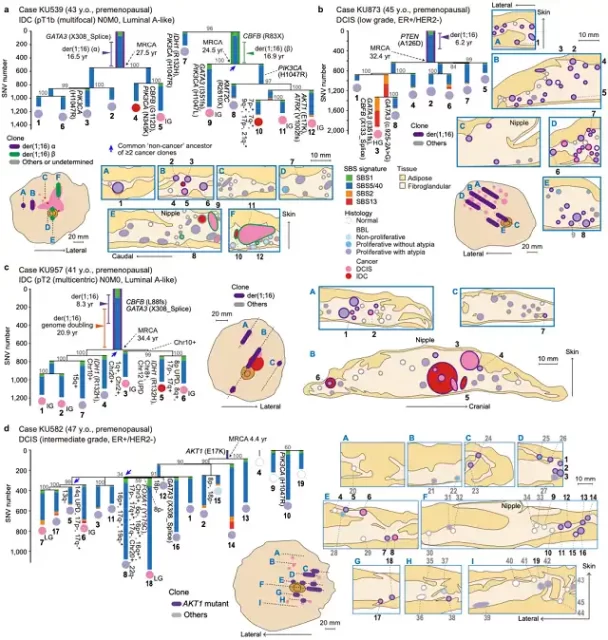
Study data for four other patients
Based on the above research data, Professor Ogawa’s team estimated the earliest occurrence time of driver mutations.
The results of their evaluation showed that the mean age of the five patients was 10.6 (5.8-16.9) years when der(1;16) appeared and 26.5 (18.1-34.4) years when MRCA appeared .
Specifically, in the case of 48-year-old Ms. KU779, two different der(1;16) were detected in her tissue samples, which were estimated to have appeared at the age of 5.8 and 10.0, and then at the age of 18.1 and 22.3, The first two clones each produced their own MRCAs .
It is worth noting that, from the above two figures, we can also see that a tumor tissue may have evolved from multiple independent ancestors at different time points . This should also be a major reason why cancers show heterogeneity.
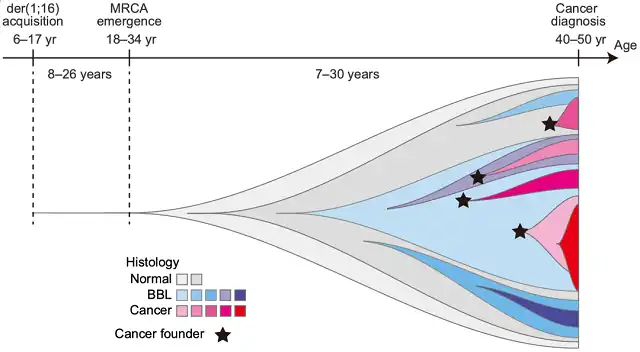
Schematic diagram of breast cancer evolution
At the end of the study, Professor Ogawa’s team described the clinical and pathological features of der(1;16)-positive breast cancer based on the data of 610 breast cancer cases published in the TCGA database, and studied der(1;16) role in breast cancer pathogenesis.
From the results of the study, der(1;16) was present in 19.5% of patients; consistent with previous studies, 86.6% of der(1;16) positive breast cancers were Luminal A type , in which der(1;16) was infiltrating The degree of enrichment was higher in infiltrating lobular carcinoma (ILC) than in invasive ductal carcinoma (IDC) (49.6% vs 12.7%).
Although the prognosis of Luminal A breast cancer is generally better, the overall survival of der(1;16) positive breast cancer is significantly longer than that of der(1;16) negative breast cancer (median OS: 33.1 months VS 28.3 months) . Even after adjusting for age and stage, der(1;16) remained a significant predictor of longer overall survival .
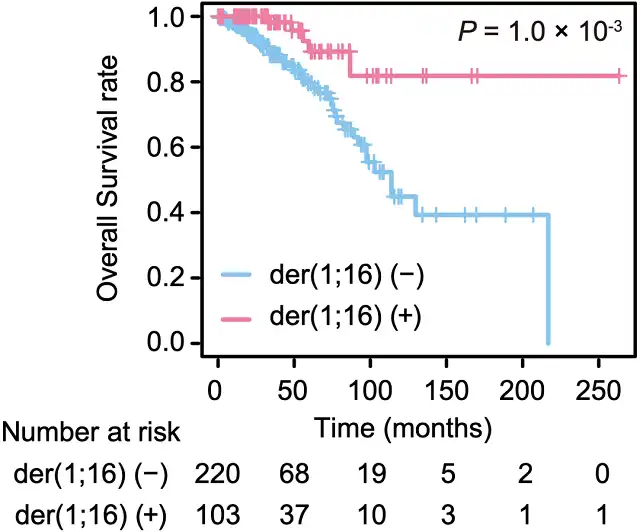
Comparison of Overall Survival
In general, the research results of the Ogawa team not only revealed the mutation process of breast epithelial cells and the life history of breast cancer, but also highlighted the unique role of der(1;16) in the main subgroup of Luminal A breast cancer , so that we have a further understanding of the occurrence and development of breast cancer.
In the future, on the basis of this research, scientists may develop markers for the prognosis of breast cancer, and also point out a new direction for the early diagnosis and even prevention of breast cancer.
references:
[1].Nishimura T, Kakiuchi N, Yoshida K, et al. Evolutionary histories of breast cancer and related clones. Nature. 2023. doi:10.1038/s41586-023-06333-9
[2].Martincorena I, Fowler JC, Wabik A, et al. Somatic mutant clones colonize the human esophagus with age. Science. 2018;362(6417):911-917. doi:10.1126/science.aau3879
[3].Yizhak K, Aguet F, Kim J, et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science. 2019;364(6444):eaaw0726. doi:10.1126/science.aaw0726
Breast cancer patients may have chromosomal mutations as early as 6 years old
(source:internet, reference only)
Disclaimer of medicaltrend.org
Important Note: The information provided is for informational purposes only and should not be considered as medical advice.



