Neglected sequences found to express universally essential proteins for cancer cell survival
- Normal Liver Cells Found to Promote Cancer Metastasis to the Liver
- Nearly 80% Complete Remission: Breakthrough in ADC Anti-Tumor Treatment
- Vaccination Against Common Diseases May Prevent Dementia!
- New Alzheimer’s Disease (AD) Diagnosis and Staging Criteria
- Breakthrough in Alzheimer’s Disease: New Nasal Spray Halts Cognitive Decline by Targeting Toxic Protein
- Can the Tap Water at the Paris Olympics be Drunk Directly?
Neglected sequences found to express universally essential proteins for cancer cell survival
Neglected sequences found to express universally essential proteins for cancer cell survival. These neglected non-coding sequences in the human genome have been found to express universally essential proteins for cancer cell survival.
Although the human genome was sequenced 20 years ago, the exact number of protein-coding genes in the human genome is still a point of dispute.
In 2001, the working draft of the Human Genome Project (HGP) was published. The project successfully mapped the human genome, and human beings obtained their own “Nature Book”. Only about 20,000 protein-coding genes were found from the human genome map. These protein-coding genes only occupy about 1.5% of the human genome, and the remaining 98.5% consist of repetitive sequences, enhancers, regulatory sequences, and non-coding RNA that are not translated These untranslated genome sequences are also called The dark genome.
However, in the next 20 years, the interpretation of this “Book of Heaven” was very difficult. Until now, we still lack sufficient understanding of the expression patterns of the human genome.
In molecular biology, open reading frames (ORFs) refer to a continuous DNA sequence starting with a start codon and ending with a stop codon, which can encode proteins. It is worth noting that genome analysis predictions indicate that there are many atypical open reading frames in the human genome, but whether they can encode biologically active proteins is not yet known.
On January 28, 2021, researchers from the Broad Institute of Massachusetts Institute of Technology-Harvard University published in Nature Biotechnology the title:
Noncanonical open reading frames encode functional proteins essential for cancer cell survival research paper.
This study shows that atypical open reading frames can also express biologically active proteins! Moreover, some atypical open reading frames are also closely related to the occurrence and development of cancer. It encodes the protein necessary for the translation of cancer cells.
In the human genome, in addition to the known 20,000 genes encoding proteins, there are also a large number of previously unknown functional proteins that play a role in diseases, especially cancer.
These proteins can be used as potential targets for cancer treatment, or will help the development of new cancer targeted therapies.
Early analysis of the human genome sequence showed that humans have 100,000 or more genes encoding proteins. However, further studies have found that most of these candidate genes are more likely to produce non-coding RNA, DNA clones with complementary fragments, or RNA with insignificant expression levels.
At the same time, the NeXtProt database of the Human Proteome Project identified about 17,600 protein-coding genes confirmed by mass spectrometry and about 2,100 unidentified protein-coding genes. However, more and more evidence using high-throughput analysis of ribosome-associated RNA indicates that DNA sequences currently annotated as non-coding RNA or pseudogenes may also encode proteins.
Moreover, due to strict, conservative analysis, only a small number of new proteins have been annotated into the human genome. It is not clear whether this translation reflects proteins that have been overlooked during the construction of reference genome databases, ribosome scans, or messy computational predictions.
More importantly, in fact, it is questioned whether these predicted new proteins can be stably translated and whether they have biological functions, which still needs to be verified by further systematic experiments.
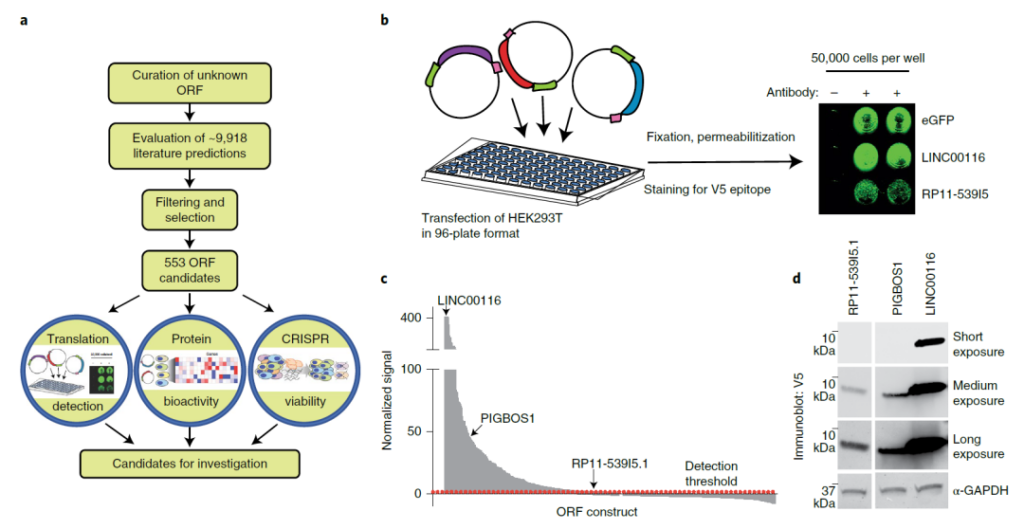
Research flow chart
In this study, the research team experimentally selected 553 candidates from the atypical open reading frame data set. The researchers found that when they were knocked out in human cancer cell lines, 57 inducible activities were lost, and after ectopic expression, 257 protein expressions and 401 induced gene expression changes.
At the same time, CRISPR screening and start codon mutation experiments show that the biological effects of atypical open reading frames are due to translation rather than RNA-mediated effects.
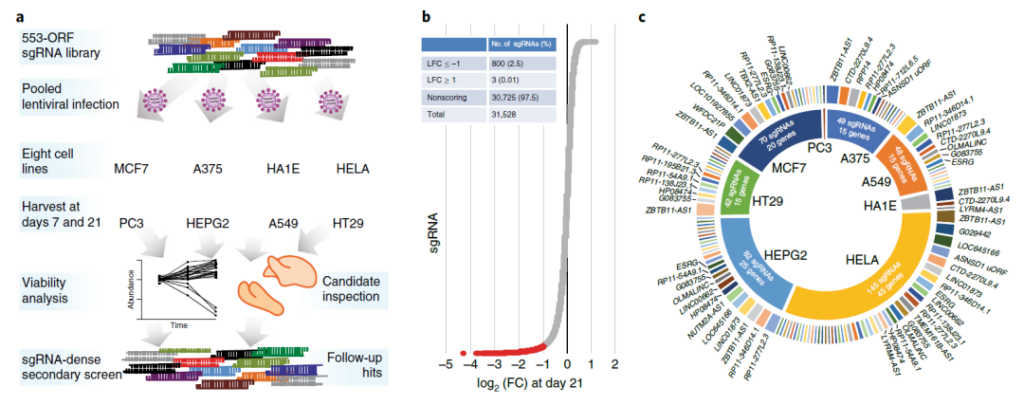
CRISPR screen to identify unknown open reading frames related to cancer cell survival
Not only that, the researchers also found one of the ORFs-glycine-rich extracellular protein 1
(GREP1), which encodes a secreted protein that is highly expressed in breast cancer. After knocking out the protein in 263 breast cancer cell lines, the proliferation of breast cancer-derived cell lines was inhibited.
In addition, studies have also shown that the abundance of GDF15, a carcinogenic factor secreted by cancer cell lines expressing GREP1, increases, and supplementation with GDF15 can alleviate cell growth inhibition after GREP1 is knocked out.
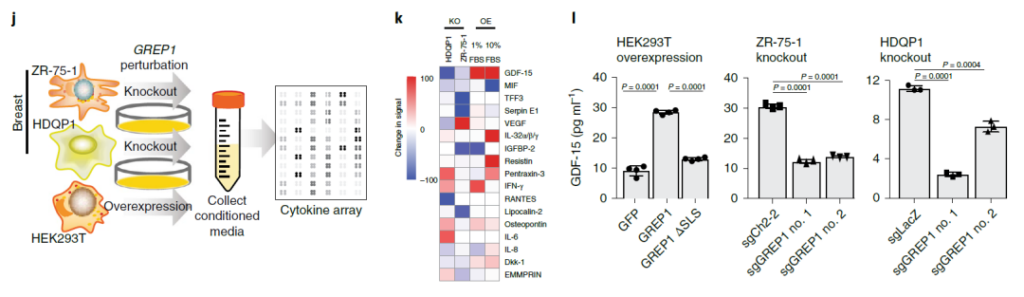
Characterization of cancer-dependent genes of GREP1 in breast cancer
The author of the paper stated: “We determined that about 10% of the open reading frames in our data set are needed for cancer cell survival. Although the data set represents a curated list of atypical open reading frames, not all possible open reading frames. Random sampling, but these experiments show that
Atypical open reading frames may provide new insights into the pathogenesis and treatment of cancer and other diseases. “
Identify translated, unannotated or unresearched atypical open reading frames
All in all, although the human genome was sequenced 20 years ago, the exact number of protein-coding genes in the human genome is still a point of dispute. This study shows that non-coding sequences outside the canonical coding region (CDS) are also commonly translated. These atypical open reading frames can also express biologically active proteins and may play a role in the occurrence and development of cancer and other diseases. Or it will become a new target for the treatment of human diseases!
(source:internet, reference only)
Disclaimer of medicaltrend.org



