The mysterious mutation in cancers: ecDNA mutation cluster is associated with 10% of cancer development
- Normal Liver Cells Found to Promote Cancer Metastasis to the Liver
- Nearly 80% Complete Remission: Breakthrough in ADC Anti-Tumor Treatment
- Vaccination Against Common Diseases May Prevent Dementia!
- New Alzheimer’s Disease (AD) Diagnosis and Staging Criteria
- Breakthrough in Alzheimer’s Disease: New Nasal Spray Halts Cognitive Decline by Targeting Toxic Protein
- Can the Tap Water at the Paris Olympics be Drunk Directly?
The mysterious mutation in cancers: ecDNA mutation cluster is associated with 10% of cancer development
- Should China be held legally responsible for the US’s $18 trillion COVID losses?
- CT Radiation Exposure Linked to Blood Cancer in Children and Adolescents
- FDA has mandated a top-level black box warning for all marketed CAR-T therapies
- Can people with high blood pressure eat peanuts?
- What is the difference between dopamine and dobutamine?
- How long can the patient live after heart stent surgery?
The mysterious mutation in cancers: ecDNA mutation cluster is associated with 10% of cancer development
It is well known that the occurrence of cancer is closely related to gene mutation. Cancer genomes contain somatic mutations that are affected by different mutational processes.
Clustered somatic mutations are common in cancer genomes, and this “clustered” refers to the concentration of these mutations in a certain part of the cancer genome. specific area.
Unfortunately, so far, aggregated somatic mutations in cancer development have been an understudied area.
But recently, Ludmil Alexandrov ‘s team at the University of California, San Diego, has discovered something very unusual about aggregated somatic mutations, and dug deeper into them.
On February 9, 2022, Ludmil Alexandrov’s team published a research paper titled: Mapping clustered mutations in cancer reveals APOBEC3 mutagenesis of ecDNA (mapping clustered mutations in cancer reveals APOBEC3 mutagenesis of ecDNA) .
This study draws the most comprehensive and detailed map of known aggregated somatic mutations , reveals the mutational signature of the cytosine deaminase APOBEC3 in extrachromatic DNA (ecDNA) , and mutates this new aggregated somatic mutation The pattern is named Kyklonas (Greek for cyclone ) , and this mutational pattern is a key previously unrecognized role in cancer evolution, responsible for the development of about 10% of human cancers and can be used to predict cancer patient survival and treatment .

Professor Ludmil Alexandrov said that normally somatic cells have random genomic mutations, but when looking closely at these mutations, they tend to occur in some hotspots .
However, clustered mutations have been largely ignored since they represent only a small percentage of all mutations. But by digging deeper, the research team found that they play an important role in the etiology of human cancers.
Previous analyses of aggregated somatic mutations have focused on single-base substitutions, revealing several types of aggregation events, including double-base substitutions (DBS) and multi-base substitutions (MBS) , omikli Mutation (Greek for mist ) and kataegis mutation (Greek for shower ) .
The Nature paper included whole-genome sequencing data of 2,583 cancer patients across 30 different cancer types , covering base substitutions and clustered insertion and deletion mutations . The research team has produced the most comprehensive and detailed map of aggregated somatic mutations known .
The research team developed next-generation artificial intelligence methods to create this map, and they then used these algorithms to detect clustered mutations in individual patients and elucidate the underlying mutational processes that lead to these events.
Through these, they found that in about 10 percent of human cancers, aggregated somatic mutations contribute to cancer development .
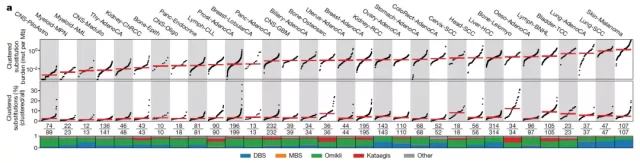 Distribution of clustered mutations in human cancers
Distribution of clustered mutations in human cancers
Not only that, but the researchers also found that some groups of cancer driver genes—especially those known to drive cancer—can be used to predict overall patient survival.
For example, the presence of clustered mutations in the BRAF gene , the most widely observed driver gene in melanoma, is more beneficial for overall patient survival than individuals with non-clustered mutations. In contrast, clustered mutations in the EGFR gene , the most widely observed driver gene in lung cancer, resulted in decreased patient survival.
Erik Bergstrom , Ph.D., the paper’s first author , said: “This is a very interesting phenomenon.
We see that different clustered mutations can produce different survival rates, and these genes are all clinically detectable. Therefore, this is a very simple and precise biomarker that can predict patient survival.
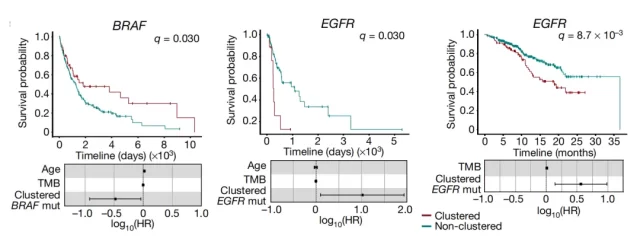
Clustered mutations in the BRAF gene improve patient survival, while clustered mutations in the EGFR gene reduce patient survival
The research team further analyzed various factors that cause aggregated somatic mutations, including ultraviolet radiation, alcohol consumption, smoking, most notably the activity of an antiviral enzyme called APOBEC3.
The APOBEC3 enzyme is normally found inside cells as part of the intracellular immune response. Their main job is to destroy viruses that enter cells.
But now, Ludmil Alexandrov’s team believes that in cancer cells, the APOBEC3 enzyme may do more harm than good. The research team found that cancer cells have looped structures of extrachromosomal DNA (ecRNA) that contain known cancer driver genes, and that there are often numerous mutations within a single ecDNA molecule.
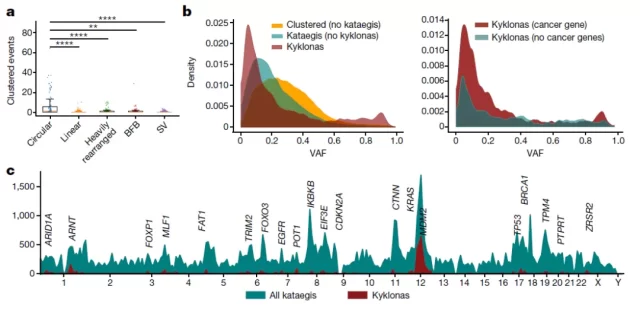
Numerous mutations in a single ecDNA molecule
The researchers attributed the mutations to the activity of the APOBEC3 enzyme, which mistook circular ecDNAs for foreign viruses and tried to restrict and cut them off .
During this process, the APOBEC3 enzyme leads to the formation of clusters of mutations within a single ecDNA molecule, which in turn plays a key role in accelerating cancer evolution and potentially leading to drug resistance. The researchers named these aggregated mutated ecDNAs ” Kyklonas ,” which means ” cyclone ” in Greek .
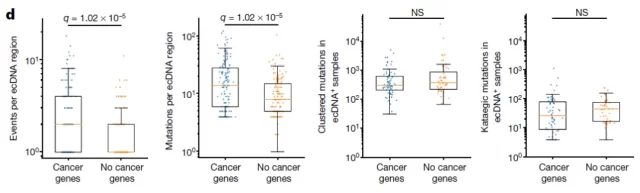
The APOBEC3 enzyme leads to the formation of clusters of mutations within a single ecDNA molecule
Professor Ludmil Alexandrov said that this is a completely new mode of tumorigenesis. More importantly, this lays the foundation for new cancer treatments that, in the future, clinicians may consider restricting the activity of the APOBEC3 enzyme or targeting ecDNA to treat cancer.
Altogether, this study provides the most comprehensive and detailed map of known aggregated somatic mutations and reveals that these aggregated mutations can be used to predict patient survival.
In addition, this study also shows that APOBEC3 enzymes and ecDNA play a key role in cancer evolution and drug resistance, providing new ideas for cancer treatment.
Paper link :
https://www.nature.com/articles/s41586-022-04398-6
The mysterious mutation in cancers: ecDNA mutation cluster is associated with 10% of cancer development
(source:internet, reference only)
Disclaimer of medicaltrend.org
Important Note: The information provided is for informational purposes only and should not be considered as medical advice.



