Why does Ebola virus become the ‘Replication Machine’?
- Normal Liver Cells Found to Promote Cancer Metastasis to the Liver
- Nearly 80% Complete Remission: Breakthrough in ADC Anti-Tumor Treatment
- Vaccination Against Common Diseases May Prevent Dementia!
- New Alzheimer’s Disease (AD) Diagnosis and Staging Criteria
- Breakthrough in Alzheimer’s Disease: New Nasal Spray Halts Cognitive Decline by Targeting Toxic Protein
- Can the Tap Water at the Paris Olympics be Drunk Directly?
Why does Ebola virus become the ‘Replication Machine’?
- Should China be held legally responsible for the US’s $18 trillion COVID losses?
- CT Radiation Exposure Linked to Blood Cancer in Children and Adolescents
- FDA has mandated a top-level black box warning for all marketed CAR-T therapies
- Can people with high blood pressure eat peanuts?
- What is the difference between dopamine and dobutamine?
- How long can the patient live after heart stent surgery?
Uncovering Ebola’s ‘replication machine’Why does Ebola virus become the ‘Replication Machine’?
In September this year, a new Ebola outbreak emerged in Uganda, Africa. This means that the Ebola virus, the “Sword of Damocles”, is still hanging overhead, threatening people’s health at all times.
The scientific community has always wanted to develop a broad-spectrum anti-Ebola virus small molecule drug that is low-cost and conducive to promotion, but lacks theoretical guidance. Ebola virus polymerase is responsible for viral genome replication and is highly conserved, making it an important target for the development of broad-spectrum drugs.
However, the molecular weight of Ebola virus polymerase is large, unstable and easy to degrade, and the analysis of its three-dimensional structure has always been a worldwide problem.
Now, the team of Academician Gao Fu from the Institute of Microbiology, Chinese Academy of Sciences and Shi Yi’s team have worked together to analyze the three-dimensional structure of the Ebola virus polymerase complex for the first time, laying a key theoretical foundation for understanding the Ebola virus replication mechanism at the molecular level.
They also analyzed the molecular mechanism by which the old drug suramin can effectively inhibit the activity of Ebola virus polymerase, providing new targets and directions for the development of anti-Ebola virus drugs. Related research was published in the journal Nature on September 28.
Structurally stable “replication machines”
Ebola virus disease is a severe acute infectious disease caused by the Ebola virus, mainly transmitted between humans or primates.
Since it was first discovered in Zaire and Sudan in 1976, the Ebola virus has been raging in Africa for nearly 50 years, with more than 30 outbreaks, causing tens of thousands of deaths, with a case fatality rate of up to 90%.
Among them, the Ebola outbreak that broke out in West African countries from 2014 to 2016 was the most serious one in history, causing a total of about 29,000 infections and more than 11,000 deaths. The 2018-2020 Ebola outbreak in the Democratic Republic of Congo also resulted in 3,481 infections and 2,299 deaths.
In 2021, active signs of Ebola virus were found again in the West African country of Guinea, suggesting that the virus may have latent infection.
These data show that scientific research on Ebola virus still needs to be strengthened.
At present, there are two Zaire Ebola virus antibody drugs on the market, the target of which is the Ebola virus surface spike glycoprotein (GP).
“The marketed antibody drugs are specific and cannot be used to treat other types of filoviruses such as Ebola virus and Marburg virus. In addition, the GP protein on the surface of the virus is also prone to immune escape under host immune pressure, which may cause Antibody drugs become less effective or even ineffective,” Shi said to “Science China Journal”, the corresponding author of the paper.
In addition, he said that due to the relatively high manufacturing cost of antibody drugs and the need for low-temperature storage, it is not conducive to popularization and use in Africa.
The development of effective and safe broad-spectrum antiviral small molecule drugs is an important research direction to deal with different types of Ebola virus and other filovirus infections.
Scientists have discovered that the transcription and replication of the Ebola virus genome is performed by a complex formed by the viral polymerase protein L and other accessory proteins.
Since the polymerase complex is highly conserved among different filoviruses, it is an ideal target for the development of broad-spectrum antiviral drugs.
Previous studies have also found that Remdesivir and Favipiravir, two nucleoside small molecule drugs targeting Ebola virus polymerase, have good in vitro antiviral activity.
However, its clinical effect is not ideal. Currently, there are no clinically approved small-molecule drugs for the treatment of Ebola virus infection.
So, how can existing targeted polymerase drugs be structurally optimized? Can new drugs be developed targeting conserved sites of polymerases to treat Ebola virus disease?
Answering these questions urges researchers to figure out how the Ebola virus replicates.
Ebola virus belongs to the Filoviridae family, and its genome is a non-segmented negative-strand RNA, about 19 kb in length, containing seven open reading frames.
In the past nearly 10 years, scientists have gained some understanding of the complex structure of this virus, such as its RNA genome is wrapped by nucleoprotein (NP) to form a ribonucleoprotein complex (RNP), which further interacts with polymerase protein (L). , viral accessory protein (VP35), transcriptional activator protein (VP30) and nucleocapsid-associated protein (VP24) to form a helical nucleocapsid structure, which is surrounded by matrix protein (VP40), and further forms a complete GP protein with the virus surface virus particles.
Deciphering the three-dimensional structure of the Ebola virus polymerase is a challenge for virologists worldwide. After years of trying and groping, the collaborative research team of Shi Yi and Gao Fu successfully expressed and purified the Ebola virus polymerase complex protein (L-VP35 complex), and used cryo-electron microscopy to analyze its high-resolution three-dimensional structure. They found that the polymerase L protein of Ebola virus forms a stable complex with the VP35 protein tetramer for viral genome replication and transcription.
So, how does the L-VP35 polymerase complex mediate this function? The research team explored this question in depth at the atomic level.
Into the Micro: Uncovering the Mechanism of “Dancing Together”
Through in-depth observations, the research team gained a microscopic level understanding of the dynamic conformational changes of the polymerase complex.
As the core of the viral “replication machine”, the polymerase involves a variety of conformational changes in the process of generating progeny RNA, thereby promoting the smooth synthesis of the product.
The most important conformational change is the transition from the initial state to the extended state.
Previous studies have shown that when the polymerase enters the extended state from the starting state, two key structural elements in the enzyme activity center, the priming loop and the supporting helix, must undergo huge conformational adjustments to ensure The product chain has enough room to extend. Otherwise, steric hindrance will occur with the product chain.
By changing the sample preparation conditions for cryo-electron microscopy, the researchers captured the fine structure of the Ebola virus polymerase in its extended state.
They found that in the extended conformation, the initiation loop is fully retracted into the capping domain, and the support helix is also farther away from the polymerase’s active center, providing sufficient space for the template/product double-stranded RNA to extend.
So, how does the viral VP35 protein “dance” with L polymerase?
The study found that in the process of viral replication, VP35 acts like a “bridge” to connect the L protein and the viral RNP. “When L polymerase replicates the viral genome, it uses helical RNP as a template instead of naked RNA.
At this time, VP35 mainly functions as a molecular chaperone, mediating L polymerase to carry out RNP as a unit. Copy.” Shi Yi said.
He explained to the “Chinese Journal of Science” that in addition to the oligomerization domain in the middle, the VP35 tetramer has four N-terminal (amino-terminal) domains and four C-terminal (carboxy-terminal) domains at both ends, one of which is C-terminal.
The end is bound to the L protein, which further stabilizes the binding of L polymerase to the VP35 tetramer, while the other seven ends will act like “octopus antennae” to help L polymerase to slide on the RNP structure, and to bind single The viral RNP protein in the body state prevents it from non-specifically interacting with the host RNA and ensures that the monomeric NP protein can be used for the generation of progeny RNP.
“Without VP35, the L polymerase protein would not be able to carry out genome replication and transcription.” Shi Yi said, if the binding of L protein and VP35 can be blocked, the virus will not be able to replicate.
Insights: Guiding Drug Design and Optimization
The development of small molecule drugs that can effectively inhibit Ebola virus has always been an international hotspot and a difficulty.
Shi Yi said that understanding the molecular details of the interaction interface of the L-VP35 complex provides new targets and important guiding information for the further development of drugs targeting polymerases.
According to reports, the Ebola virus polymerase activity domain and capping domain are structurally similar to other (genome) unsegmented negative-strand RNA virus polymerases such as respiratory syncytial virus (RSV) and rabies virus (RABV). It shows that this kind of viral polymerase is conserved in the evolution process.
Notably, the researchers pointed out that the N-terminal domain region of Ebola virus polymerase has a filovirus-specific insertion domain and is essential for Ebola virus polymerase activity. as a potential target for antiviral drug development.
In this study, the research team also explored the molecular mechanism of the in vitro anti-Ebola virus activity of the century-old drug suramin.
Suramin is an odorless, tasteless white powder first isolated by the German chemist Paul Ehrlich in the early 20th century, soluble in physiological saline, and later found to be useful in the treatment of various parasitic diseases, from It was widely used in the early 1920s to treat parasitic diseases such as African sleeping sickness and onchocerciasis.
In recent years, scientists have found that suramin has anti-coronavirus and cancer activity. Preliminary studies suggest that suramin also has anti-Ebola virus activity, but its mechanism of action is unclear.
Through in vitro enzyme activity and cellular replicon experiments, the researchers found that suramin can effectively inhibit the activity of Ebola virus polymerase, and further analyzed the complex structure of Ebola virus polymerase and suramin using cryo-electron microscopy. , revealing that suramin exerts its inhibitory effect by binding to the NTP entry channel of the polymerase and preventing the substrate from entering the enzyme activity center.
The molecular details of the interaction between suramin drugs and L protein provide key reference information for further engineering and optimization of suramin drugs.
“Suramin is a potential antiviral drug, and a highlight of this study is the dissection of the L protein-suramin complex, which can guide further antiviral drug design,” said a reviewer of the paper. . A
nother peer reviewer stated that the lack of structural information on the Ebola virus L protein has been a well-recognized gap in the field, and this study provides important information that could help facilitate structure-based antiviral drug design.
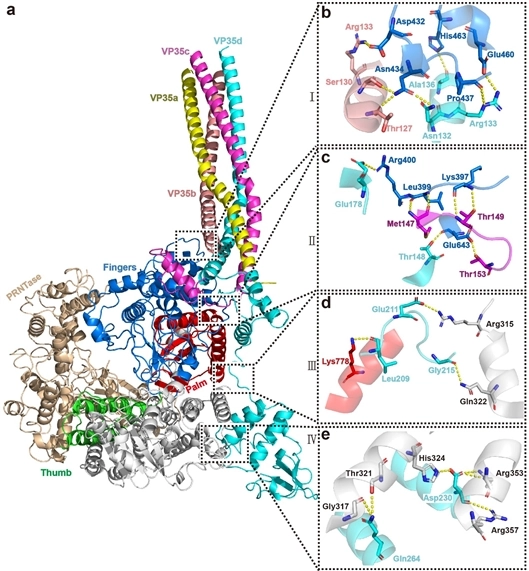
The details of the interaction between L protein and VP35 are provided by the author
Related paper information:
https://doi.org/10.1038/s41586-022-05271-2
Why does Ebola virus become the ‘Replication Machine’?
(source:internet, reference only)
Disclaimer of medicaltrend.org
Important Note: The information provided is for informational purposes only and should not be considered as medical advice.



