Alcohol causes cancer depending on whether you have such mutated gene
- Normal Liver Cells Found to Promote Cancer Metastasis to the Liver
- Nearly 80% Complete Remission: Breakthrough in ADC Anti-Tumor Treatment
- Vaccination Against Common Diseases May Prevent Dementia!
- New Alzheimer’s Disease (AD) Diagnosis and Staging Criteria
- Breakthrough in Alzheimer’s Disease: New Nasal Spray Halts Cognitive Decline by Targeting Toxic Protein
- Can the Tap Water at the Paris Olympics be Drunk Directly?
- Should China be held legally responsible for the US’s $18 trillion COVID losses?
- CT Radiation Exposure Linked to Blood Cancer in Children and Adolescents
- FDA has mandated a top-level black box warning for all marketed CAR-T therapies
- Can people with high blood pressure eat peanuts?
- What is the difference between dopamine and dobutamine?
- How long can the patient live after heart stent surgery?
Alcohol causes cancer depending on whether you have such mutated gene.
The Lancet Oncology: Drinking alcohol is easy to get liver cancer? See if you have this mutation! The first GWAS of alcohol-related liver cancer patients discovered a pair of important cancer-related genes
Have you ever had a similar myth: some friends around you, stay up late for liver essays, fried chicken, milk tea, happy water, and stay alive after only 5 hours of sleep a day, while yourself, if you sleep an hour late, the next day The brain is blank, the heart rate is too fast, the eyes are dull, and the hair keeps losing…
Although it must be admitted that it is true that living habits are not good, why are some people so “made”?
For example, drinking alcohol is not good for the liver. This is a common sense, but in fact, a considerable number of people who have been drinking for a long time have not developed liver cancer.
In a recent article published in The Lancet Oncology, scientists from France and Belgium conducted a genome-wide association study (GWAS) to investigate the pathogenesis associated with alcohol-related hepatocellular carcinoma (HCC). The role of genes in population inheritance.
They identified a pair of genes, WNT3A-WNT9A, that may be associated with alcohol-related HCC, which may play an early role in the Wnt-β-catenin pathway associated with the development of liver cancer. This is also the first GWAS performed in this type of patient [1].

Alcohol-related liver disease accounts for nearly one-third of HCC patients worldwide [2].
Some clinical studies have found that the risk factors associated with alcohol consumption that may lead to HCC include advanced age, male sex, liver fibrosis, etc.
However, some people who drink alcohol for a long time and occupy the above risk factors do not progress to HCC [2-4], indicating that in addition to the environment Factors, individual genetic differences also play a decisive role.
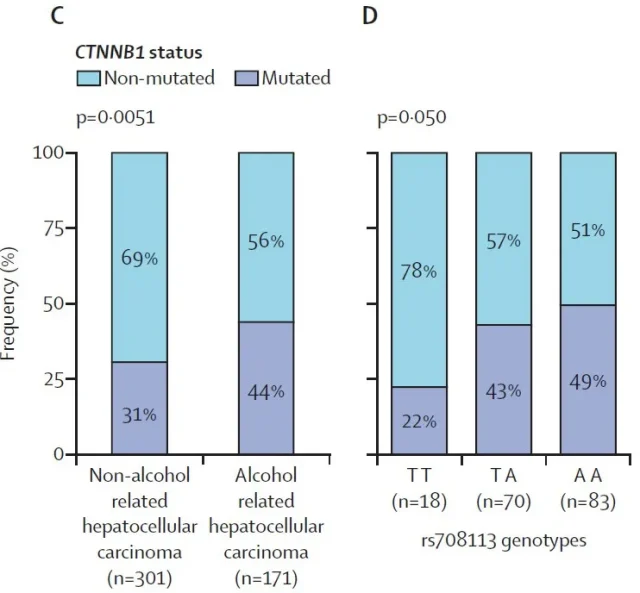
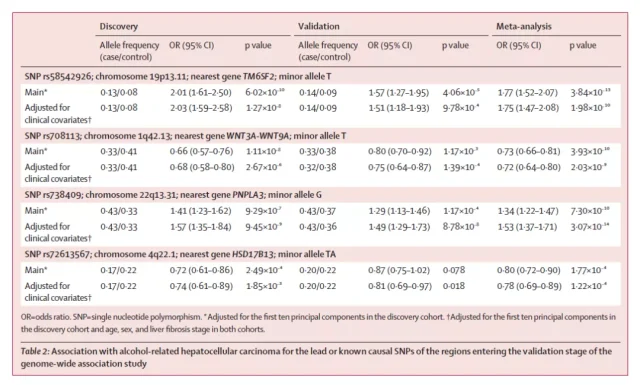
Past GWAS studies have identified two common single nucleotide base mutations associated with increased risk of HCC: rs738409(C>G) in PNPLA3 and rs58542926(C>T) in TM6SF2 [5,6], and One associated with reduced risk of HCC, rs72613567(T>TA) in HSD17B13 [7].
In this study, a GWAS study was conducted for these common single nucleotide base mutations found to help us better understand the impact of these genes in the pathogenesis of alcohol-related HCC and hepatocellular carcinoma.
The subjects were unrelated European patients with alcohol-related liver disease and were divided into 3 phases: discovery cohort, validation cohort, and meta-analysis.
In the discovery cohort, the researchers analyzed samples from 775 HCC patients (case group) and 1332 non-HCC liver disease patients (non-case group), and the results showed that rs8107974 in the SUGP1 gene was in high linkage discordance with rs5852926 in TM6SF2. Balanced state, which means that the two base pairs 7974 and 2926 are more likely to be inherited together.
Similarly, rs2294915 and rs738409 in PNPLA3 and rs708113 in WNT3A-WNT9A were also in high linkage disequilibrium with two other single-nucleotide base pairs in the same region.
The gene HSD17B13, which is associated with reduced HCC risk, was also validated in the discovery cohort, but did not reach GWAS-level significance.
Notably, in the discovery cohort, the researchers identified a third gene associated with an increased risk of HCC: WNT3A-WNT9A.
They were also confirmed in the validation cohort that these three genes were indeed associated with an increased risk of HCC.
The researchers also investigated the association between the number of genetic mutations and the risk of alcohol-related HCC in a meta-analysis.
According to the analysis results, the number of mutations carried and the incidence of alcohol-related HCC have an additive effect , that is, the more mutations carried, the greater the risk of alcohol-related HCC.
This was true even after adjusting for confounding factors such as age, sex, and stage of liver fibrosis.
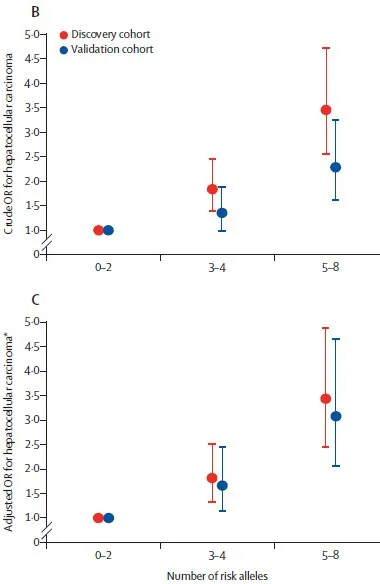
HCC risk in carriers of different numbers of risk alleles
Overall, the above three-phase analysis first identified four gene mutations (three increases and one decrease) associated with alcohol-related HCC risk, but the effects of mutations associated with reduced risk were not significant enough. In addition, the more mutations carried, the higher the risk of developing HCC, regardless of age, gender and other factors.
In addition, in order to compare with other types of chronic liver disease (chronic hepatitis C, chronic hepatitis B, non-alcoholic fatty liver disease), the researchers also conducted a GWAS study in patients with the above three chronic liver diseases.
They found that rs708113 in the WNT3A-WNT9A gene was not associated with the risk of developing HCC for these three chronic liver diseases, while the other three mutations were significantly associated with the risk of developing HCC from nonalcoholic fatty liver disease.
The researchers extracted non-tumor tissue from patients with alcohol-related HCC and found that mutations in the WNT3A-WNT9A rs708113[T] base pair were associated with high expression of inflammatory factors and natural killer cells.
At the same time, they also studied the difference in the expression level of this gene between alcohol-related HCC patients and non-alcohol-related HCC patients.
Before that, we briefly understand a gene mutation commonly found in alcohol-related HCC patients, CTNNB1 gene mutation, which is associated with activation of the Wnt-β-catenin pathway and low immune infiltration in the tumor environment. All are related to the occurrence of HCC [8,9].
The researchers found that patients with alcohol-related HCC had more CTNNB1 gene mutations than non-alcohol-related HCC patients, but in patients with alcohol-related HCC, patients with WNT3A-WNT9A rs708113 mutation had decreased CTNNB1 gene mutation.
The “seesaw” balance of mutations in these two genes was associated with the amount of alcohol consumed , and the researchers found their association only in HCC patients who drank alcohol.
So far, this study has demonstrated the association of common gene mutations found with alcohol-related HCC at the gene level, and completed the discovery and validation of WNT3A-WNT9A rs708113.
Since WNT3A and WNT9A encode ligands for the Wnt-β-catenin pathway [10], which plays an important role in the development, metabolism, and hepatocarcinogenesis of liver disease, mutations in this pathway may seriously affect regulation Gene signal transduction.
Alcohol-related HCC usually has a poor prognosis, and these findings affirm the important role of the WNT3A-WNT9A locus in the pathogenesis of alcohol-related HCC and may be a potential target in the future treatment of alcohol-related HCC.
However, whether the rs708113 mutation has an effect on the expression of WNT3A-WNT9A in tumor tissue, or whether it directly affects the tumor microenvironment, still needs to be explored.
references:
1. Eric Trépo, et al. Common genetic variation in alcohol-related hepatocellular carcinoma: a case-control genome-wide association study, The Lancet Oncology, 2021,
2. Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2021; 7: 6.
3. Yu MW, Chang HC, Liaw YF, et al. Familial risk of hepatocellular carcinoma among chronic hepatitis B carriers and their relatives. J Natl Cancer Inst 2000; 92: 1159–64.
4. Hemminki K, Li X. Familial liver and gall bladder cancer: a nationwide epidemiological study from Sweden. Gut 2003; 52: 592–96.
5. Trépo E, Nahon P, Bontempi G, et al. Association between the PNPLA3 (rs738409 C>G) variant and hepatocellular carcinoma: evidence from a meta-analysis of individual participant data. Hepatology 2014; 59: 2170–77.
6 Yang J, Trépo E, Nahon P, et al. PNPLA3 and TM6SF2 variants as risk factors of hepatocellular carcinoma across various etiologies and severity of underlying liver diseases. Int J Cancer 2019; 144: 533–44.
7. Abul-Husn NS, Cheng X, Li AH, et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med 2018; 378: 1096–106.
8. Schulze K, Imbeaud S, Letouzé E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015; 47: 505–11.
9. Ruiz de Galarreta M, Bresnahan E, Molina-Sánchez P, et al. β-catenin activation promotes immune escape and resistance to anti-PD-1 therapy in hepatocellular carcinoma. Cancer Discov 2019; 9: 1124–41.
10. Parsons MJ, Tammela T, Dow LE. WNT as a driver and dependency in Cancer. Cancer Discov 2021; 11: 2413–29.
Alcohol causes cancer depending on whether you have such mutated gene
(source:internet, reference only)
Disclaimer of medicaltrend.org
Important Note: The information provided is for informational purposes only and should not be considered as medical advice.