Antibody screening technology in innovative antibody discovery
- Why Lecanemab’s Adoption Faces an Uphill Battle in US?
- Yogurt and High LDL Cholesterol: Can You Still Enjoy It?
- WHO Releases Global Influenza Vaccine Market Study in 2024
- HIV Infections Linked to Unlicensed Spa’s Vampire Facial Treatments
- A Single US$2.15-Million Injection to Block 90% of Cancer Cell Formation
- WIV: Prevention of New Disease X and Investigation of the Origin of COVID-19
Application of antibody screening technology in innovative antibody discovery
- Red Yeast Rice Scare Grips Japan: Over 114 Hospitalized and 5 Deaths
- Long COVID Brain Fog: Blood-Brain Barrier Damage and Persistent Inflammation
- FDA has mandated a top-level black box warning for all marketed CAR-T therapies
- Can people with high blood pressure eat peanuts?
- What is the difference between dopamine and dobutamine?
- What is the difference between Atorvastatin and Rosuvastatin?
- How long can the patient live after heart stent surgery?
Application of antibody screening technology in innovative antibody discovery.
Antibody discovery includes antibody screening and antibody optimization.
This article makes a detailed comparison of the technical procedures and application characteristics of hybridomas, phage display antibody libraries, cell-free molecular display antibody libraries, cell surface antibody libraries, and B cell gene cloning technologies.
Mouse hybridoma and phage display antibody libraries are widely used in antibody screening due to their technological maturity and fluency.
The cell-free expression display antibody library is suitable for introducing mutations in vitro and adding screening pressure; the cell surface display antibody library can be used to finely and highly enrich high-affinity antibodies due to the use of flow sorting technology.
Therefore, these two technologies have unique advantages in antibody optimization.
In contrast, B-cell gene cloning technology uses high-throughput single-cell operations (including single-cell separation, screening, PCR, and sequencing), which requires higher equipment, reagents, consumables, and operating technology, and the application cost is also the highest.
In addition, it is necessary to accumulate more single-cell screening experience and methods for individual screening of diversified targets, and it will take time for the promotion of antibody screening in the future.
Antibody discovery includes two processes: antibody screening and optimization.
Antibody screening is to search for high-quality candidate antibodies from the beginning, and antibody optimization is to make necessary improvements to existing antibodies.
Antibody discovery is the only way to develop monoclonal antibody drugs, bispecific antibody drugs, ADC drugs and Car-T cell therapy drugs.
According to the clonal selection theory, animal immunity enriches dominant B cell clones in vivo; while in vitro affinity panning or cell sorting (Sorting) enriches dominant antibody clones in vitro.
Natural immunogens (such as viruses or cells) are easier to enrich in vivo B cell clones that recognize natural epitopes than artificially prepared immunogens (such as polypeptides or recombinant proteins); if artificially prepared immunogens are used to immunize animals, they will inevitably Generate B cell clones that are enriched to recognize unnatural epitopes because unnatural epitopes are more immunogenic.
The key indicator for screening functional antibodies is more than antibody affinity.
Affinity panning or cell sorting based on antigen/antibody interaction should avoid over-enrichment of high-affinity antibodies, and should take into account the diversity of antibody affinity and antibody sequence, and avoid losing rare functional antibody clones.
Appropriately reducing the screening pressure and expanding the screening throughput will help improve the success rate of screening functional antibodies.
Hybridomas and antibody libraries are the most widely used antibody screening technologies.
After continuous improvement of the gene cloning technology of a single B cell, it has also begun to be used for antibody screening. Hybridomas include mouse, rat, hamster and rabbit hybridoma technology; antibody libraries include phage display antibody libraries, cell-free molecule display antibody libraries (ribosome display, mRNA display and DNA display antibody libraries), and cell surface display antibody libraries (Bacterial surface display, fungal surface display, yeast surface display and mammalian cell surface display antibody library).
Compares the operating procedures of various antibody screening technologies.
1. Hybridoma
So far, hybridoma technology has developed very mature. The so-called hybridoma is not only a hybrid cell selected in a limited medium after fusion of B cells with antibody secretion ability and infinite proliferation ability of myeloma cells in vitro, it has both antibody secretion ability and infinite in vitro proliferation ability.
Hybridomas have only the in vivo enrichment process produced by immunity, and there is no in vitro enrichment process.
During screening, the culture supernatant of hybridoma cells can be directly detected to complete the preliminary screening.
Mouse hybridomas can directly prepare mouse ascites purified monoclonal antibodies; rat, hamster and rabbit hybridomas cannot prepare ascites, and hybridoma cells need to be cultured in vitro to purify monoclonal antibodies; all species of hybridomas can be cloned or cloned from the initial screening.
The antibody gene is cloned in the subcloned cells, molecular transformation is carried out, and the recombinant monoclonal antibody is expressed and purified. After the monoclonal antibody is purified, the functional antibody can be screened and identified through meticulous functional evaluation.
The efficiency of cell electrofusion to prepare hybridomas is very high, and the B cells of each mouse can produce up to 2-4×10^4 candidate hybridoma clones.
However, since each hybridoma cell is the product of the fusion of B cells and myeloma cells, it is unstable during cell division and easily loses the ability to produce antibodies.
Therefore, it must be subcloned to isolate stable antibody-secreting hybridomas. clone. During subcloning, it is easy for the positive clones to turn negative in the initial screening.
Therefore, completing gene cloning before subcloning can avoid losing the antibody genes of the positive clones in the initial screening.
The time window before subcloning is very short and the number of cells is small, which limits the flux of gene cloning and molecular conversion before subcloning and reduces the successful screening rate of functional antibodies.
Therefore, the use of hybridoma approach to screen functional antibodies usually requires immunization of multiple animals, and sometimes repeated immunizations due to fusion or screening failure.
This is especially true when developing antibody drugs against difficult targets.
2. Antibody Library
2.1 Types of antibody libraries
According to the source and use of antibody genes, antibody libraries are divided into two categories: immune antibody libraries and general antibody libraries.
The antibody genes of the immune antibody library are derived from immunized human or animal B cells (lymph nodes, spleen and peripheral blood lymphocytes).
The general antibody library is divided into natural antibody library, fully synthetic antibody library and semi-synthetic antibody library.
According to the different forms of antibody molecules, the antibody library can be divided into three main types: Fab antibody library, scFv antibody library and VHH antibody library.
The VHH antibody library is also called Nano antibody library.
The antibody genes of the natural antibody library are derived from unimmunized human or animal B cells.
According to the clonal selection theory of antibody production, after the fetal organs mature, clonal selection will bring about the bias of B cell cloning.
There are a large number of B1 and B2 cell clones that have not been cloned in fetal liver and bone marrow, and the diversity of antibodies is higher. , So it is the best choice for building a natural antibody library.
Since B cells in lymph nodes, spleen and peripheral blood lymphocytes have undergone in vivo clonal selection, more donors need to be collected to cover as much antibody diversity as possible.
According to the principle of antibody gene rearrangement, the V and D interfaces of the variable region of the antibody heavy chain, the D gene fragment, and the D and J interfaces together constitute the heavy chain CDR3, which is also HCDR3; the V and J interfaces of the light chain variable region are It constitutes the light chain CDR3, which is LCDR3.
The combined diversity of different V, D, and J gene clusters, the uncertainty at the interface during gene rearrangement, and the somatic mutations that cover the variable region genes of the antibody, plus the combined diversity of heavy and light chains, are common It constitutes the entire source of antibody diversity.
In the interaction between antigen and antibody, the variable region of the heavy chain is more important than the variable region of the light chain, and the diversity of HCDR3 is the highest, so HCDR3 contributes the most.
Figure 2 shows the corresponding relationship between the gene rearrangement of the antibody variable region, the primary sequence of the antibody variable region and the structure of the antibody.
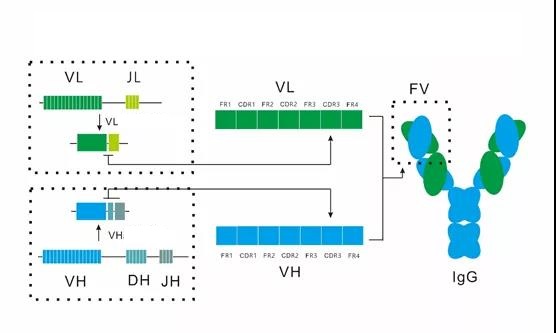 Figure 2. The corresponding relationship between antibody variable region gene rearrangement, primary sequence of antibody variable region and antibody structure
Figure 2. The corresponding relationship between antibody variable region gene rearrangement, primary sequence of antibody variable region and antibody structure
Based on the detailed analysis of antibody sequence and structure, the antibody heavy chain variable region and light chain variable region were divided into 7 groups.
Combine the most frequently occurring amino acids in each position of the variable region into one sequence, that is, consensus sequence.
Using bioinformatics methods, the amino acids at each position in all the 6 CDR regions of the antibody variable region were predicted, and 6 CDR peptide libraries were designed.
The consensus sequence of the heavy chain and the light chain is randomly combined to generate 49 sets of backbone sequences, and then embedded with six CDR peptide libraries to design scFv or Fab antibody libraries.
This is the design principle of Morphosys’ fully human fully synthetic antibody library (HuCAL) . On this basis, after this series of optimization operations, such as adjusting the gene sequence according to the host cell codon bias, deeply optimizing the amino acid composition and ratio of the CDR peptide library, according to the natural antibody heavy and light chain variable region pairing bias, artificial design Determine the combination tendency of the heavy chain and light chain variable region skeleton sequence, remove the specific amino acid clusters that affect the physical and chemical properties of the antibody protein, and the design of the fully synthetic antibody library is also continuously upgraded.
Natural antibody libraries and fully synthetic antibody libraries have their own advantages and disadvantages.
The antibody genes of the natural antibody library are derived from natural B cells, and the diversity of epitopes is relatively limited, which also affects the affinity of the antibodies to be screened.
However, due to the in vivo evolution process, indicators such as stability, solubility and expression levels perform better.
In contrast, the design diversity of the fully synthetic antibody library reaches more than 10^20. Even if the bacterial transformation step is restricted during the construction of the antibody library, the diversity of antibody recognition epitopes far exceeds that of the natural antibody library.
However, this kind of artificially designed sequence diversity has not undergone in vivo evolution. It often occurs with abnormal protein modification or abnormal amino acid clusters, low expression levels and easy degradation.
Therefore, the antibodies screened out need to be optimized for the necessary antibodies to be more patented. Sex.
The semi-synthetic antibody library is designed to integrate the advantages of the natural antibody library and the fully synthetic antibody library, and try to overcome their shortcomings.
The FR1-CDR1-FR2-CDR2-FR3 gene fragments of the antibody variable region are all derived from natural B cells, while the CDR3 region and FR4 are artificially synthesized.
The amino acid composition and ratio of each position in the CDR3 region are also designed by bioinformatics technology.
Therefore, the semi-synthetic antibody library not only reduces the proportion of artificial design, but also ensures sufficient antibody diversity.
2.2 Comparison of antibody libraries based on different display technologies
According to different display technologies, antibody libraries are divided into three categories: phage display antibody libraries, cell-free molecule display antibody libraries, and cell surface display antibody libraries.
The cell-free molecular display antibody library can be divided into three types: Ribosome display, mRNA display and DNA display antibody library. According to different host cells, the cell surface display antibody library can be divided into four types: bacterial surface display, fungal surface display, yeast surface display, and mammalian cell surface display antibody library.
Due to the limitations of transformation efficiency, cell culture density and cell culture volume, the upper limit of the storage capacity of the cell surface display antibody library (especially the mammalian cell surface display antibody library) is relatively small (generally less than 10^9). During screening, single cell clones are usually enriched and separated by cell sorting, followed by high-throughput single-cell PCR to clone antibody genes and perform molecular transformation, and then express and purify recombinant antibodies for screening and Identify functional antibodies.
Because gene cloning and molecular conversion are both limited by operating throughput, the throughput of candidate recombinant antibody expression, purification and functional screening is limited. The lower the throughput, the easier it is to lose high-quality clones.
Among the various cell surface display antibody libraries, yeast and mammalian cells have more mature protein synthesis mechanisms, which are more conducive to displaying antibodies that are close to their natural conformations. The binding activity of antibodies screened from the corresponding cell surface display antibody libraries is better.
Reflects the activity of natural antibodies. Since the upper limit of the storage capacity of the cell surface display antibody library is relatively small, and the cell sorting is easy to enrich high-affinity antibodies, the cell surface display antibody library is more suitable for antibody optimization work such as affinity maturation.
The construction of the cell-free molecular display antibody library only requires the preparation of the corresponding DNA template in vitro, and the step of transforming bacteria or cells is completely unnecessary, so the upper limit of the storage capacity is the largest, even greater than 10^16.
The antibody library stored in the form of DNA needs to undergo PCR amplification, in vitro transcription and in vitro translation operations before screening to realize the correlation between antibody genotype (RNA or DNA) and phenotype (protein).
Affinity panning is usually used to complete in vitro enrichment during screening.
After polyclonal PCR amplification and polyclonal molecular conversion, bacteria are transformed and single clones are picked, and prokaryotic cells or mammalian cells are used as the The host expresses the recombinant antibody. Finally, the unpurified antibody is used for preliminary screening, and the purified antibody is used to complete the screening and identification of functional antibodies.
Since the affinity panning step is completely completed in a cell-free in vitro environment, it is easier to add screening pressure (such as temperature, pH, salt concentration, detergent concentration, etc.), so it has a unique optimization in the physical and chemical properties of antibodies Advantage.
The step of polyclonal PCR will obviously increase the copy number of the monoclonal, which requires subsequent molecular conversion, antibody expression and purification, and antibody screening with higher throughput to obtain higher antibody sequence diversity.
Due to the smaller size of the bacterial host and phage particles, the large culture density and small volume, and the efficiency of bacterial transformation is also higher than that of yeast and mammalian cells, the limit storage capacity of the phage display antibody library is between the cell surface display antibody library and the cell-free molecular display Among antibody libraries, the upper limit is generally 10^11-10^12, which is sufficient to meet the application requirements of immune antibody libraries and general antibodies.
The phage display antibody library is also enriched in vitro by affinity panning, and then the activity of the phage display antibody or prokaryotic secreted and expressed antibody fragments is detected to complete the preliminary screening. Phage particles allow long-term storage and facilitate high-throughput screening.
And the molecular conversion link is simple and easy. Compared with the other two types of antibody library screening technologies, the advantages of the phage display antibody library are summarized as follows: large storage capacity, simple operation, low cost, suitable for high throughput, and beneficial to the screening of functional antibodies.
Based on this, the phage display antibody library has developed into one of the mainstream techniques for antibody screening.
3. Comparison of antibody library and hybridoma
As a commonly used antibody screening technology, hybridoma and phage display antibody libraries have their own characteristics and advantages.
The heavy chain and light chain of hybridoma monoclonal antibodies are derived from a B cell, which is a natural pairing.
The antibody library is the product of a random combination of heavy and light chains from different B cells.
On the one hand, the unnaturally paired heavy and light chains may affect the activity of the antibody, but on the other hand, it also increases the diversity of the antibody library and helps optimize the activity of the antibody.
Even for hybridoma monoclonal antibodies, in the antibody optimization stage, chain replacement strategies are often used to optimize the pairing combination of light and heavy chains, improve antibody affinity and optimize other functions of the antibody.
Hybridoma and phage display antibody libraries are used in antibody discovery, requiring users to have different technical reserves.
Mouse hybridomas do not need to go through the steps of gene cloning, molecular conversion and expression of recombinant antibodies.
They only need to go through animal immunization, cell culture and cell fusion, mouse ascites preparation and monoclonal antibody purification to complete the preliminary screening and functional identification of antibodies.
The technical requirements are relatively simple. Rats, hamsters, and rabbits hybridomas cannot prepare ascites, or they can be cultured in vitro to purify the antibodies, or they can be cloned and molecularly transformed to express and purify recombinant antibodies. This increases the technical difficulty and complexity as well as The screening throughput is limited, and high-quality clones are easily lost. In comparison, in addition to animal immunity and immunoassays, phage display antibodies are completely genetic engineering technology systems, including library construction, molecular conversion, and expression and purification of recombinant antibodies.
Therefore, the promotion and application of phage display antibody libraries are closely related to the development and maturity of genetic engineering antibody technology.
The antibody library is a combination of a full set of antibody gene cloning technology and antibody surface display technology.
When using antibody libraries, animal immunization, antibody library construction, and antibody screening are independent links.
After immunization, it is sufficient to retain B cells or nucleic acids for future library construction, without the need to retain animals or living cells; once the antibody library is built, it can be stored permanently, without repeated immunization and library building, allowing repeated screening; Each antibody gene is cloned individually and directly undergoes molecular transformation.
These are the main advantages that distinguish antibody libraries from hybridomas.
Candidate clones of hybridomas cannot be stable for a long time, and the time window for primary screening and subcloning is very short. Sometimes repeated immunization, fusion and screening are required, which takes time and effort.
The broad species spectrum of the antibody library is also its advantage. Its antibody genes are not only derived from mouse, rat, hamster and rabbit animal species that can produce hybridomas, but also include humans, non-human primates, camels, Alpacas, sharks, chickens and other species that cannot perform effective cell fusion due to lack of suitable myeloma cell lines.
In theory, only need to use transcriptome sequencing and other technical means to obtain the antibody gene sequence, then primers can be designed to clone the full set of antibody genes of the species, and an antibody library can also be constructed to screen antibodies.
4. Single B cell cloning technology
Essentially a single B cell antibody gene cloning technology is a B cell surface display technology. Natural B cells cannot survive and expand indefinitely in vitro. Only when single cell isolation, single cell screening, single cell PCR or single cell sequencing technologies are used in a comprehensive manner, can high-throughput preliminary screening and obtaining different B cells in a short time window. Antibody genes cloned by cells.
Only with high-throughput molecular conversion can detailed evaluation and identification and screening of functional antibodies be completed.
The key here is how to design a high-throughput single-cell screening method according to the requirements of screening functional antibodies to reduce the scale and throughput of candidate operations.
Berkeley Light launched the Beacon device to solve high-throughput single-cell separation and single-cell screening, but the equipment is too expensive, the cost of consumables is too high, and the single-cell screening methods and strategies are too simple to be enriched.
At present, this technology is widely used in the screening of antibodies against viral infectious diseases (such as the screening of new coronavirus neutralizing antibodies), and the promotion and application of relatively complex functional antibody screening in the future has yet to be tested in practice.
5. Antibody optimization
Antibody discovery also includes antibody optimization areas such as humanization, affinity maturation, and optimization of physical and chemical properties. Antibody optimization is a kind of molecular directed evolution.
First construct a variety of mutant antibody libraries (including phage display antibody libraries, cell surface display antibody libraries or cell-free molecule display antibody libraries), and then use affinity panning, cell sorting, or other biological, physical or chemical indicators based Special screening methods to screen and optimize certain characteristics of antibodies.
The design and construction of mutant antibody libraries can use random mutation (DNA shuffling or error-prone PCR), site-directed mutation or local region mutation, and chain shuffling (chain shuffling) and other mutation strategies and technical means.
As mentioned earlier, the cell-free molecular display technology is not only suitable for introducing mutations at the molecular level, but also convenient for adding different screening pressures in the screening process based on optimization indicators.
The cell surface display mostly uses cell sorting strategies to enrich high-affinity clones in vitro, which is more suitable for antibody affinity maturation. In practice, you can choose from three different antibody libraries according to the design capacity of the mutant antibody library.
The storage capacity of cell-free molecule display is the largest (the upper limit is 10^16), and the storage capacity of phage display antibody is in the middle (the upper limit is 10 ^10-10^12), the cell surface display antibody library has the smallest storage capacity (upper limit is less than 10^9).
6. Conclusion
In short, antibody discovery is a comprehensive technical system including animal immunity, molecular cloning, immunological testing, protein production, cell function evaluation and animal testing.
Among various screening technologies, phage display antibody libraries and hybridomas have their own characteristics and advantages, and are the most widely used.
Other screening technologies have also flourished because of their uniqueness.
In the process of antibody drug development, antibody discovery requires in-depth research on the individuality of each target.
According to the specific application requirements of disease treatment, and under the premise of grasping the scientific nature of the target, various antibody screening technologies can be rationally used to develop antibody drugs. Provide high-quality candidate functional antibodies.
Application of antibody screening technology in innovative antibody discovery
(source:internet, reference only)
Disclaimer of medicaltrend.org



